Int. J. Dev. Biol. 67: 9 - 17 (2023)
Circ-JA760602 promotes the apoptosis of hypoxia-induced cardiomyocytes by transcriptionally suppressing BCL2
Original Article | Published: 5 April 2023
Abstract
Acute myocardial infarction (AMI) is myocardial necrosis caused by the complete or partial obstruction of a coronary artery. Circular RNAs (circRNAs) have been proven as regulators in the progression of various human diseases, including AMI. However, the role of novel circ-JA760602 in AMI remains unknown. Here, we investigated the role of circ-JA760602 in modulating the apoptosis of hypoxia-induced AMI cells using the AC16 cardiomyocyte in vitro cell model. The expression of circ-JA760602 in AC16 cardiomyocytes subjected to hypoxia was measured by quantitative real-time polymerase chain reaction (qRT-PCR). Cell viability was measured by cell counting kit-8 (CCK-8) assay. Apoptosis of cardiomyocytes was evaluated by TUNEL assay and flow cytometry analysis. The cellular location of circ-JA760602 was identified through fluorescence in situ hybridization (FISH) assay and subcellular fractionation assay. The downstream molecular mechanisms of circ-JA760602 were demonstrated by luciferase reporter assay, RNA binding protein immunoprecipitation (RIP) assay and chromatin immunoprecipitation (ChIP) assay. Rescue assays were performed to demonstrate the effects of BCL2 knockdown on circ-JA760602 silencing-mediated cardiomyocyte apoptosis. Circ-JA760602 expression was elevated after hypoxia treatment. Knockdown of circ-JA760602 enhanced viability and curbed apoptosis of hypoxia-treated cardiomyocytes. EGR1 and E2F1 could activate BCL2 transcription. Cytoplasmic circ-JA760602 bound with EGR1 and E2F1 to thus inhibit their nuclear translocation. BCL2 knockdown reversed the effects of circ-JA760602 silencing on the apoptosis of hypoxia-treated AC16 cells. Circ-JA760602 promotes hypoxia-induced apoptosis of cardiomyocytes by binding with EGR1 and E2F1 to inhibit the transcriptional activation of BCL2.
Keywords
circ-JA760602, BCL2, apoptosis, acute myocardial infarction
Introduction
Cardiovascular diseases (CVDs) have become a major threat to people’s health worldwide. Great efforts have been made to improve diagnosis and treatment of CVDs, including medical imaging, bioengineering, animal and clinical studies (Tang et al., 2015). Acute myocardial infarction (AMI) mainly refers to myocardial necrosis caused by acute and persistent ischemia and hypoxia of the coronary artery (Feng et al., 2021). AMI accounts for 81% of cardiogenic shock cases (Kapur et al., 2020) and leads to nearly half of death cases of CVDs (Feng et al., 2021). Currently, reperfusion of infarcted coronary arteries is considered as the most effective treatment method for AMI, which is used for restoration of oxygen supply and blood flow (Wang et al., 2021a). Although reperfusion can help patients survive from AMI, the injury after treatment still causes much pain to patients (Kloner et al., 2016). Consequently, it is of great importance explore the molecular mechanisms involved in AMI for the better prognosis of AMI patients.
Circular RNAs (circRNAs) are a class of endogenous non-coding RNAs with stable covalent closed loop structure, which can involve in the development of varied diseases, including AMI. For instance, circMACF1 has been found to impede AMI progression via miR-500b-5p/EMP1 axis (Zhao et al., 2021). CircROBO2 has been proven to promote AMI via targeting miR-1184/TRADD axis (Chen et al., 2021). In addition, circUBXN7 can alleviate the hypoxia/reoxygenation-induced cell apoptosis and inflammation in AMI (Wang et al., 2021a). In this study, circ-JA760602 (hsa_circ_0089761) was chosen as the research object due to its up-regulation in heart samples of AMI patients, according to GEO dataset. Considering that AMI can result in apoptosis and injury of cardiomyocytes (Jiao et al., 2019; Wu et al., 2017), we intended to figure out whether circ-JA760602 can regulate the apoptosis of cardiomyocytes in AMI model.
In this study, we constructed an AMI cell model through treating AC16 cells with hypoxia and explored the mechanism by which circ-JA760602 affected hypoxia-induced apoptosis of AC16 cells. By means of this study we hope to provide a better understanding of the mechanism underlying the progression of AMI.
Results
Circ-JA760602 expression is upregulated in hypoxia-induced cardiomyocytes
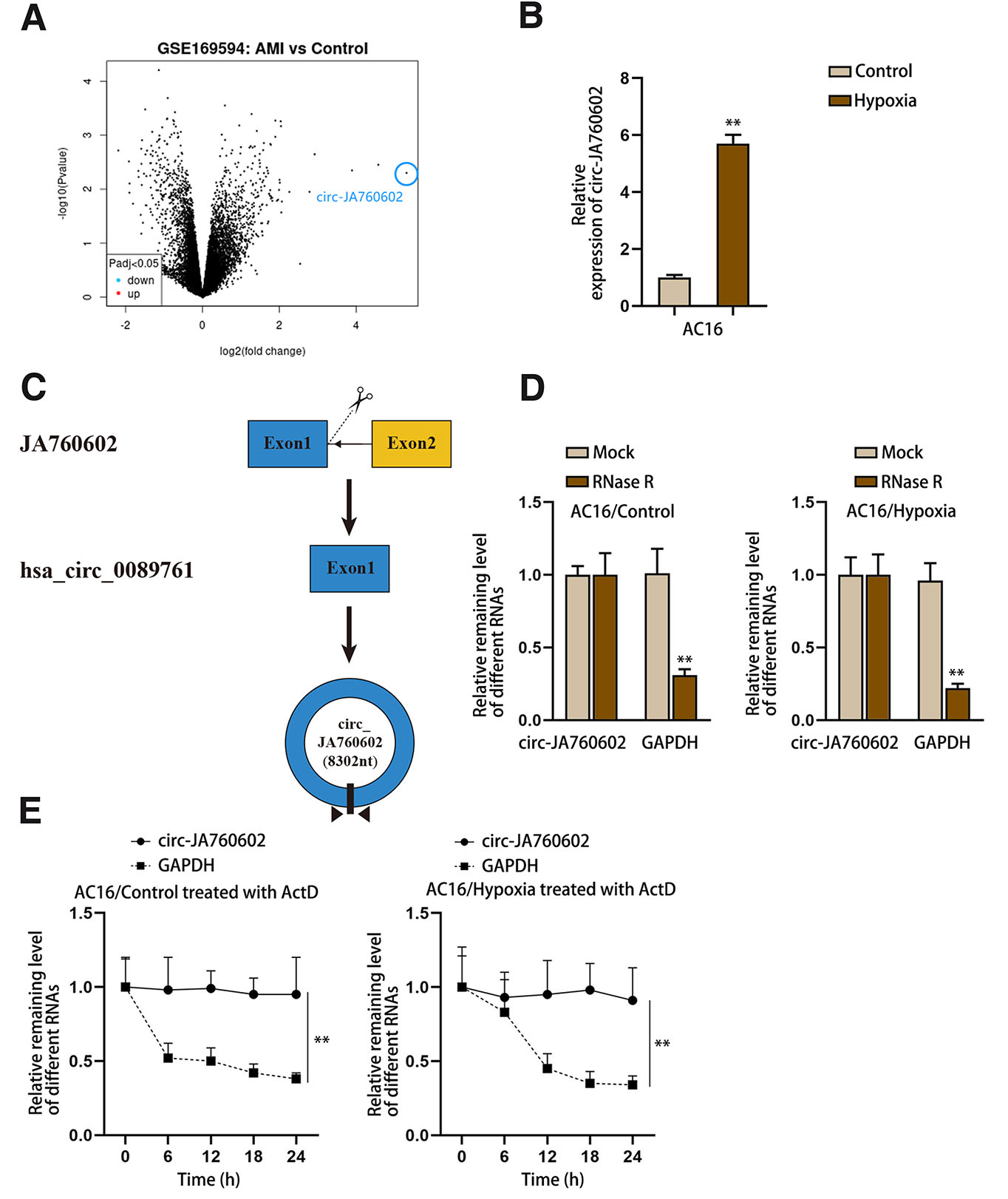
Through analyzing the GEO dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169594), we determined that circ-JA760602 was most significantly upregulated in the heart samples collected from AMI patients (Fig. 1A). Next, we uncovered that circ-JA760602 expression was significantly augmented in AC16 cells after hypoxia treatment through qRT-PCR measurement (Fig. 1B). Therefore, we chose circ-JA760602 as the research object. Before further detection, we demonstrated the circular characteristics of circ-JA760602. As shown in Fig. 1C, circ-JA760602 was generated from exons 1 of its host gene JA760602. Then, we detected the stability of it in cells treated with RNase R or actinomycin D (ActD). It was found that GAPDH mRNA was digested by RNase R, while circ-JA760602 was resistant to RNase R digestion (Fig. 1D). Moreover, circ-JA760602 degraded by actinomycin D (ActD) was slower than GAPDH mRNA (Fig. 1E). Thus, we confirmed that circ-JA760602 is a bona fide circRNA. To conclude, circ-JA760602 expression was elevated in cardiomyocytes upon hypoxia treatment.
Fig. 1. Circ-JA760602 is highly expressed in hypoxia-induced cardiomyocytes.
(A) CircRNAs, significantly up-regulated in human acute myocardial infarction (AMI) tissues compared with control tissues, were screened out from GSE169594 dataset. (B) qRT-PCR assay detected the expression of circ-JA760602 in AC16 cells treated with or without hypoxia. (C) Schematic of the circular structure of circ-JA760602. (D,E) qRT-PCR assay examined the expression of circ-JA760602 in normoxic and hypoxic AC16 cells treated with RNase R (D) or ActD (E). GAPDH was used as the linear control. **P < 0.01.
Circ-JA760602 knockdown suppresses the apoptosis of hypoxia-induced cardiomyocytes
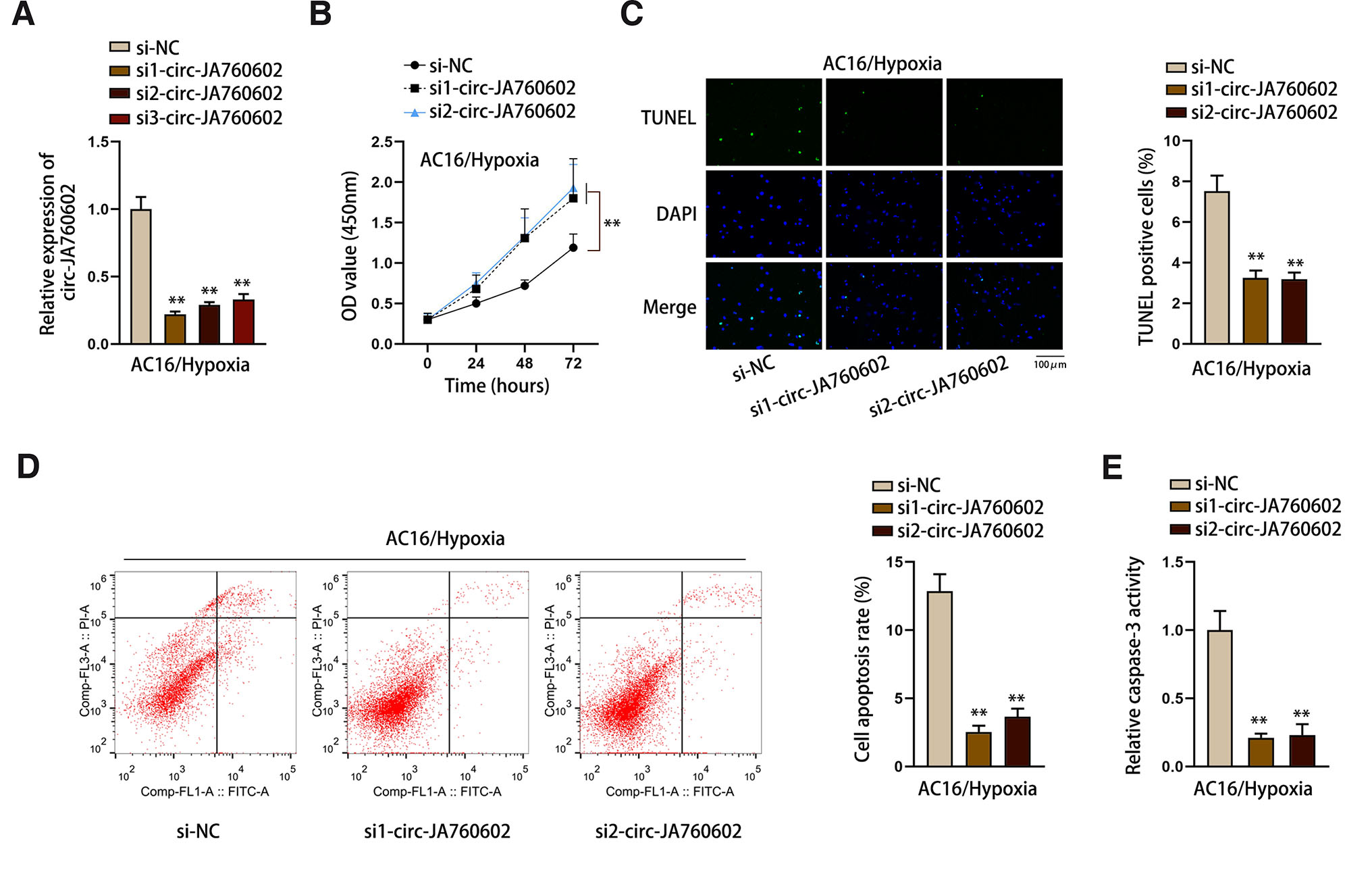
To determine the functional role of circ-JA760602 in hypoxia-induced AMI cell model, we first knocked down circ-JA760602 expression by transfecting circ-JA760602-specific siRNAs into Hypoxia-AC16 cells (Fig. 2A). We selected si1-circ-JA760602 and si2-circ-JA760602 for the loss-of-function assays due to their high knockdown efficiencies. The results of CCK-8 assay indicated that cell viability was notably promoted after knockdown of circ-JA760602 (Fig. 2B). Moreover, cell apoptosis was markedly inhibited after circ-JA760602 silencing, as demonstrated by the decreased TUNEL positive cell number and cell apoptosis rate (Fig. 2 C-D). Meanwhile, the caspase-3 activity was weakened when circ-JA760602 expression was down-regulated, also suggesting that cell apoptosis was hindered (Fig. 2E). Taken together, circ-JA760602 down-regulation could suppress hypoxia-induced apoptosis of cardiomyocytes.
Fig. 2. Circ-JA760602 knockdown suppresses the apoptosis of hypoxia-induced cardiomyocytes.
(A) qRT-PCR assay tested the expression of circ-JA760602 in hypoxia-induced AC16 cells after the transfection of circ-JA760602-specific siRNAs. (B) CCK-8 assay detected viability of hypoxic AC16 cells upon circ-JA760602 deficiency. (C-E) TUNEL, flow cytometry, and caspase-3 activity assays were applied to evaluate TUNEL positive cell percent (C), cell apoptosis rate (D), and relative caspase-3 activity (E) in hypoxia-treated AC16 cells after circ-JA760602 knockdown. **P < 0.01.
Circ-JA760602 modulates BCL2 transcription via EGR1 and E2F1
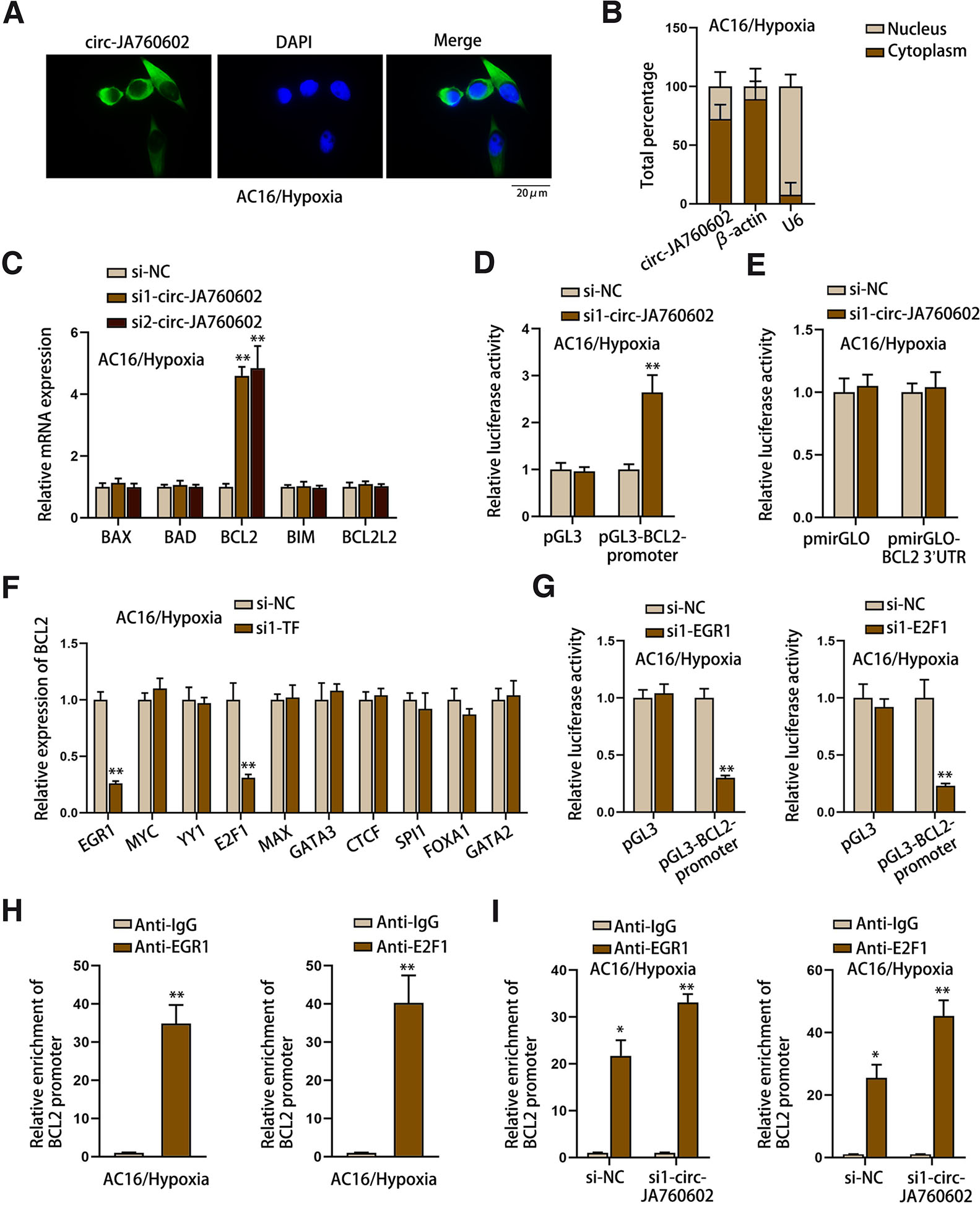
Thereafter, we continued to explore the regulatory mechanism of circ-JA760602. The subcellular location of circ-JA760602 was first analyzed by FISH and subcellular fractionation assays. As displayed in Fig. 3 A-B, circ-JA760602 mainly distributed in the cytoplasm of hypoxia-induced AC16 cells. Next, we detected the expression of apoptosis-related genes in circ-JA760602-silenced cells using qRT-PCR. As exhibited in Fig. 3C, only BCL2 was significantly upregulated upon circ-JA760602 depletion. To investigate the interaction between circ-JA760602 and BCL2, we conducted luciferase reporter assays with pGL3-BCL2-promoter or pmirGLO-BCL2 3’UTR. As shown in Fig. 3D, the luciferase activity of pGL3-BCL2-promoter was notably enhanced by circ-JA760602 knockdown. However, the luciferase activity of pmirGLO-BCL2 3’UTR was almost unchanged after knockdown of circ-JA760602 (Fig. 3E). These data indicated that circ-JA760602 regulated BCL2 transcription. To investigate the specific regulatory mechanism, we searched on UCSC (http://www.genome.ucsc.edu) for the potential transcription factor of BCL2. Among the predicted transcription factors, E2F1 (Ma et al., 2021), MYC (Yu et al., 2018), YY1 (Bhaskar Rao et al., 2021), EGR1 (Liu et al., 2018), MAX (Lafita-Navarro et al., 2020), GATA3 (Qi et al., 2021), CTCF (Peng et al., 2019), SPI1 (Luo and Ge, 2020), FOXA1 (Zhang et al., 2020), GATA2 (Cao et al., 2022a) have been proven to promote the transcription of genes. For further screening, we knocked down these transcription factors and then detected the expression of BCL2. As a result, BCL2 expression was significantly downregulated only when EGR1 or E2F1 expression was knocked down (Fig. 3F). Then, luciferase reporter assay reflected that the luciferase activity of pGL3-BCL2-promoter was weakened after silencing of EGR1 or E2F1 (Fig. 3G). ChIP assay further validated the binding of EGR1 and E2F1 to BCL2 promoter (Fig. 3H). Moreover, we found that circ-JA760602 inhibited the affinity of EGR1 and E2F1 to BCL2 promoter (Fig. 3I). Hence, it could be concluded that circ-JA760602 negatively regulates BCL2 via affecting the EGR1/E2F1-mediated transcription.
Fig. 03. Circ-JA760602 depletion enhances the transcriptional activity of BCL2.
(A,B) FISH and subcellular fractionation assays determined the distribution of circ-JA760602 in hypoxia-induced AC16 cells. (C) qRT-PCR assay detected the expressions of apoptosis-related genes (BAX, BAD, BCL2, BIM and BCL2L2) in AC16 cells under hypoxia. (D,E) Luciferase reporter assays examined the luciferase activity of BCL2 promoter and BCL2 3’UTR in hypoxia-induced AC16 cells upon circ-JA760602 depletion. (F) qRT-PCR assay detected BCL2 expression in hypoxic AC16 cells with the depletion of candidate transcription factors. (G) Luciferase reporter assay detected the luciferase activity of BCL2 promoter in EGR1/E2F1-depleted AC16 cells under hypoxia. (H) The enrichment of BCL2 promoter in the immunoprecipitates incubated by anti-EGR1 or anti-E2F1 in hypoxic AC16 cells was measured by ChIP assay. (I) ChIP assay detected the enrichment of BCL2 promoter in the immunoprecipitates incubated by anti-EGR1 or anti-E2F1 in hypoxia-induced AC16 cells upon circ-JA760602 depletion. *P < 0.05, **P < 0.01.
Circ-JA760602 binds with EGR1 and E2F1 in cytoplasm to inhibit their nuclear translocation
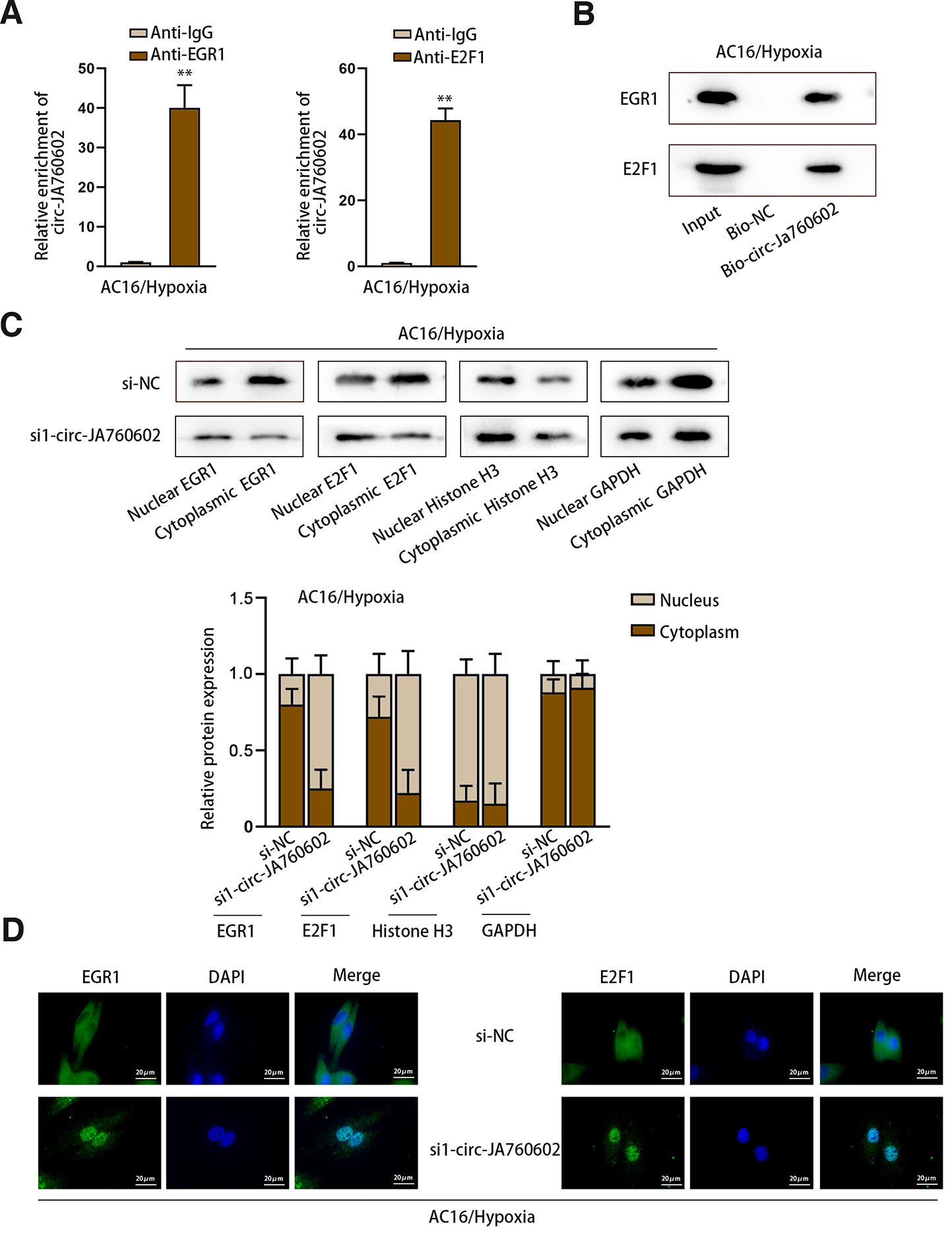
Furthermore, we explored how circ-JA760602 affected the interaction between EGR1/E2F1 and BCL2 promoter. It was found from RIP assay that circ-JA760602 could bind with EGR1 and E2F1 (Fig. 4A). RNA pull-down assay showed that EGR1 and E2F1 could be pulled down by Bio-circ-JA760602 (Fig. 4B), further confirming the interaction between EGR1/E2F1 and circ-JA760602. Then, we detected the protein levels of EGR1 and E2F1 in cell cytoplasm and nucleus after circ-JA760602 depletion. As illustrated in Fig. 4C, the nuclear distribution of EGR1 and E2F1 was increased in circ-JA760602-silenced cells. Moreover, through IF assay, we determined that knockdown of circ-JA760602 increased the nuclear accumulation of EGR1 and E2F1 (Fig. 4D). In sum, circ-JA760602 can bind to EGR1 and E2F1 to inhibit their nuclear translocation, thus suppressing BCL2 transcription.
Fig. 4. Circ-JA760602 binds with EGR1 and E2F1 to impede their nuclear translocation.
(A) RIP assay detected the enrichment of circ-JA760602 in the immunoprecipitates incubated with anti-EGR1 or anti-E2F1 in hypoxia-induced AC16 cells. (B) RNA pull-down assay measured the enrichment of EGR1 and E2F1 in the product pulled down by Bio-circ-JA760602 or Bio-NC in hypoxia-induced AC16 cells. (C,D) Subcellular fractionation-western blot and IF assays detected the location of EGR1 and E2F1 in hypoxic AC16 cells upon circ-JA760602 deficiency. **P < 0.01.
Circ-JA760602 modulates the hypoxia-induced apoptosis of cardiomyocytes via BCL2
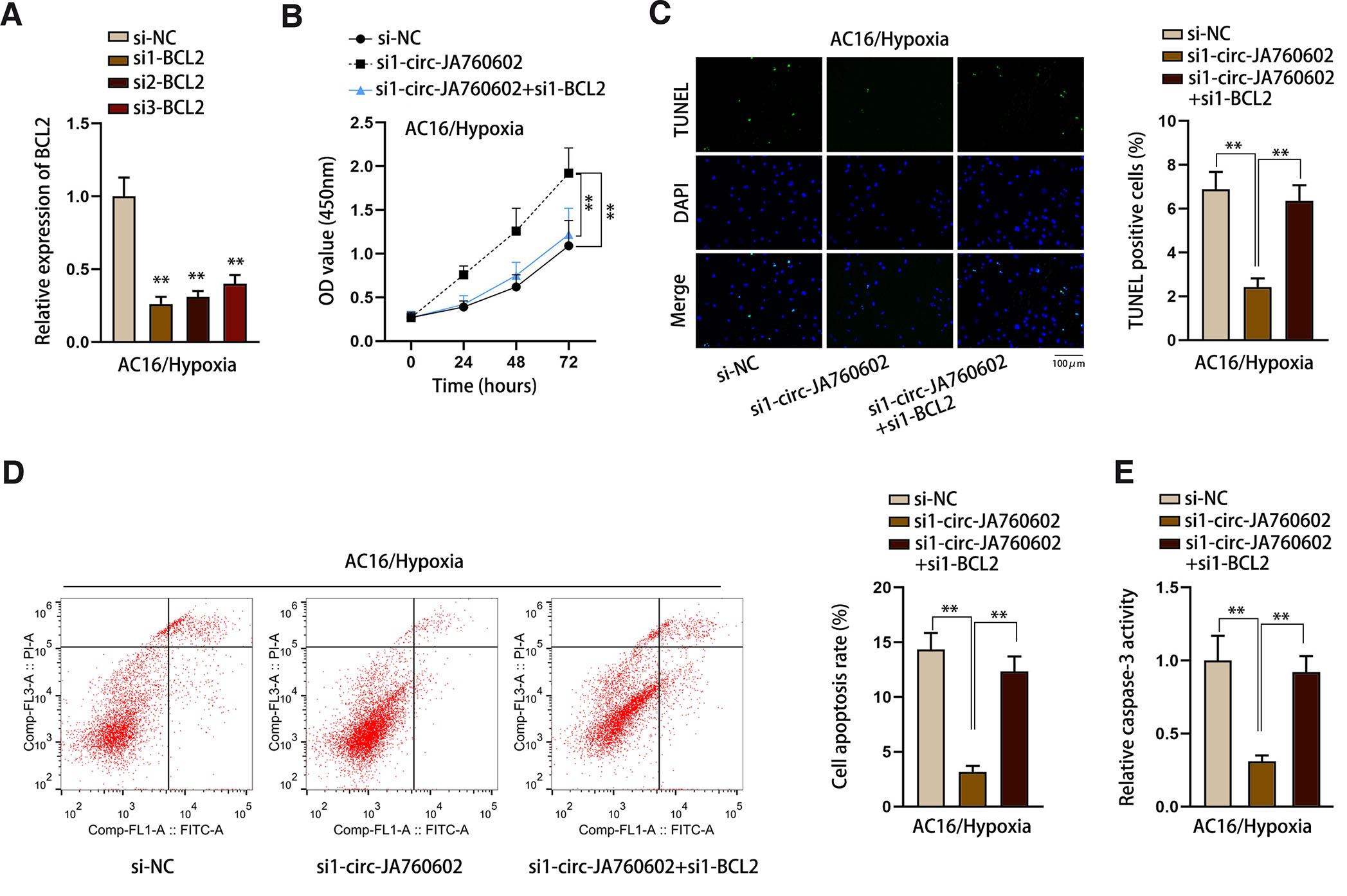
Rescue experiments were conducted to confirm that circ-JA760602 promoted hypoxia-induced cardiomyocyte apoptosis through negatively modulating BCL2. The knockdown efficiency of si1/2/3-BCL2 was firstly determined by qRT-PCR (Fig. 5A). si1-BCL2 was chosen for the subsequent rescue assays due to its highest knockdown efficiency. CCK-8 assay revealed that the enhanced viability of hypoxia-induced AC16 cells caused by circ-JA760602 knockdown was reversed by silencing of BCL2 (Fig. 5B). Importantly, it was proven that the apoptosis of hypoxia-induced AC16 cells suppressed by circ-JA760602 knockdown was rescued by BCL2 inhibition (Fig. 5 C-E). In summary, circ-JA760602 negatively modulates BCL2 expression to promote hypoxia-induced apoptosis of cardiomyocytes.
Fig. 5. Circ-JA760602 represses apoptosis of hypoxia-induced cardiomyocytes via the negative modulation of BCL2 expression.
(A) qRT-PCR assay tested BCL2 expression in hypoxia-induced AC16 cells transfected with BCL2-specific siRNAs. (B) CCK-8 assay assessed the viability of hypoxic AC16 cells subsequent to transfection with si-NC, si1-circ-JA760602 or si1-circ-JA760602+si1-BCL2. (C-E) TUNEL assay, flow cytometry analysis, and caspase-3 activity detection examined the percentage of TUNEL positive cells, cell apoptosis rate, and relative caspase-3 activity in hypoxia-induced AC16 cells under different conditions. **P < 0.01.
Discussion
Hypoxia is the major reason for ischemia, which can result in AMI (Li et al., 2020a). This study constructed an AMI cell model by treating AC16 cells with hypoxia. Moreover, this study explored a novel molecular pathway affecting hypoxia-induced apoptosis of cardiomyocytes. More and more circRNAs have been regarded as crucial regulators involved in cardiac disease (Huang et al., 2019). For instance, circRNA MFACR can facilitate hypoxia-induced cardiomyocyte apoptosis (Wang et al., 2021b). Cai et al., have reported that hypoxia induced the overexpression of circJARID2 to promote apoptosis of cardiomyocytes (Cai et al., 2021). Here, we searched on online database GEO and analyzed a dataset to select AMI-related circRNAs. According to the result, hsa_circ_0089761 (termed as circ-JA760602) was most significantly up-regulated in the heart samples of patients with AMI. Therefore, we chose it as our research object. Circ-JA760602 is a novel circRNA that was not reported in AMI. To our knowledge, the current study firstly revealed the role of circ-JA760602 in AMI-induced cardiomyocyte apoptosis. We also detected its expression in hypoxia-induced AC16 cells and the results indicated that circ-JA760602 was upregulated in AMI cell model. It is well known that circular RNAs are stable than linear RNAs (Liu and Chen, 2022). Here, we identified the circular structure of circ-JA760602 in terms of its stability. We uncovered that circ-JA760602 resisted to RNase R digestion and ActD degradation. Moreover, results of loss-of-function assay indicated that circ-JA760602 depletion enhanced the viability but decreased apoptosis rate of hypoxia-induced cardiomyocytes. Therefore, our current study firstly revealed the role of circ-JA760602 in modulating the apoptosis of hypoxia-induced cardiomyocytes.
Subsequently, we explored the downstream molecular mechanism of circ-JA760602. We firstly identified the cytoplasmic location of circ-JA760602 in hypoxia-induced AC16 cells. circRNAs can affect cell apoptosis in various disease models by regulating the expression of apoptosis-related mRNAs (Chen et al., 2021a; Yang et al., 2022; Lu et al., 2021). Considering the effect of circ-JA760602 on apoptosis, we detected its modulation on the expression of apoptosis-related mRNAs. The result revealed that circ-JA760602 knockdown led to the downregulation of BCL2, indicating that circ-JA760602 could negatively regulate BCL2 mRNA expression. Moreover, we determined that circ-JA760602 suppressed BCL2 transcription. Since circ-JA760602 mainly located in the cytoplasm, we hypothesized that it might regulate BCL2 transcription by regulating the upstream transcription factors of BCL2 in the cytoplasm. Through bioinformatics prediction and mechanism analyses, we confirmed that circ-JA760602 down-regulation could strengthen the affinity of EGR1 and E2F1 to BCL2 promoter. Hereto, we concluded that circ-JA760602 downregulated BCL2 expression by regulating EGR1 and E2F1.
Recent studies have presented that circRNAs can interact with RNA binding proteins (RBPs) to regulate the expression of their target genes (Zang et al., 2020). For example, hsa_circ_0000848 modulates the proliferation and apoptosis of hypoxia-induced cardiomyocytes through recruiting ELAVL1 and further enhancing SMAD7 stability (Cao et al., 2022b). According to the previous studies, circRNAs can affect the nuclear accumulation of proteins. For example, circ1662 can bind with YAP1 and facilitate its nuclear translocation to affect SMAD3 pathway in colorectal cancer cells (Chen et al., 2021b). CircIPO7 facilitates nuclear location of YBX1 in nasopharyngeal carcinoma cells (Hong et al., 2022). This study uncovered that circ-JA760602 could bind with EGR1 and E2F1 in the cytoplasm of cardiomyocytes. Noteworthily, circ-JA760602 down-regulation contributed to the nuclear translocation of EGR1 and E2F1, leading to the promotion of BCL2 mRNA transcription. Finally, we performed rescue assays and verified that BCL2 down-regulation abrogated the suppressive impact of circ-JA760602 knockdown on the apoptosis of hypoxia-induced cardiomyocytes.
In summary, this study firstly revealed that circ-JA760602 was upregulated in a cellular model of AMI. Concomitantly, down-regulation of circ-JA760602 impeded apoptosis of hypoxic cardiomyocytes. Mechanistically, circ-JA760602 suppressed BCL2 transcription by binding to EGR1 and E2F1 to hinder their nuclear translocation. We hope our findings have contributed to our understanding of the molecular mechanisms underlying hypoxia-induced cardiomyocyte apoptosis. Lack of in vivo and clinical data is a limitation of this study. Thus, it is our aim to establish animal models and eventually perform clinical research in our future studies.
Materials and methods
Cell culture
Human cardiomyocyte (AC16) was purchased from American Type Culture Collection (ATCC; Manassas, VA, USA), and was cultivated in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Rockford, IL, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Procell, Wuhan, China). For normoxic culture, AC16 cells were maintained in a humidified environment at 37°C with 5% CO2 and 95% air. As described previously (Yan and Hou, 2021), AC16 cells were cultured in a modular incubator with 5% CO2, 1% O2 and 94% N2 for hypoxic injury induction.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Trizol reagent (Invitrogen) was used to treat AC16 cells for extracting total RNA. Then, extracted RNAs were reversely transcribed into cDNA by using Prime Script™ RT kit (Takara, Dalian, China). The primers used in this experiment were listed in Table 1. qRT-PCR was conducted on ABI 7500 fast PCR System (Applied Biosystems, Foster city, CA, USA) with a SYBR green PCR Master Mix (Applied Biosystems). Relative RNA expression was calculated based on 2-∆∆Ct method. β-actin served as the reference gene for circRNAs and mRNAs.
Table 1
Primers used in qRT-PCR
| Genes | Sequences |
|---|---|
| circ-JA760602 (divergent primers) | F: TGAGGGCGTGATCATGAAAGG |
| R: ACCTGAATCGGAGGACAACC | |
| circ-JA760602 (convergent primers) | F: TGTTAGGGACGGATCGGAGA |
| R: TCCTAGGCGACCCAGACAAT | |
| BAX | F: ATGGACGGGTCCGGGG |
| R: TCAGCTGCCACTCGGAAAAA | |
| BAD | F: TTCTGAGGGGAGACTGAGGTCC |
| R: GATCTGGAACATGCTCTGGGC | |
| BCL2 | F: ATAACGGAGGCTGGGTAGGT |
| R: TTTATTTCGCCGGCTCCACA | |
| BIM | F: CTGAAGGCAATCACGGAGGT |
| R: CACTGGAGGATCGAGACAGC | |
| BCL2L2 | F: CTGACCCGGCTCCACG |
| R: AAGCGGGTCTCGAACTCATC | |
| β-actin | F: ACAGAGCCTCGCCTTTGCC |
| R: TGGGGTACTTCAGGGTGAGG | |
| GAPDH | F: AAGGTGAAGGTCGGAGTCAA |
| R: AATGAAGGGGTCATTGATGG | |
| U6 | F: GCTTCGGCAGCACATATACTAAAAT |
| R: CGCTTCACGAATTTGCGTGTCAT |
Cell transfection
Small interfering RNAs (siRNAs) targeting circ-JA760602 or BCL2 were designed and synthesized by Ribobio (Guangzhou, China) for knockdown of circ-JA760602 or BCL2; meanwhile, non-targeted siRNA (si-NC) was used as the corresponding negative control (NC). The constructed plasmids were transfected into AC16 cells in logarithmic growth phase using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Cells were harvested for further experiments after 48 h. Sequences for siRNAs used in this experiment was listed in Table 2.
Table 2
Sequences for siRNAs used in transfection
| siRNAs | Sequences |
|---|---|
| si-NC | 5’-GCAUGUGCCAUAGGCCCAUUU-3’ |
| si1-circ-JA760602 | 5’-UAACUAAUACUAACAUCUCAG-3’ |
| si2-circ-JA760602 | 5’-GAUGUUAGUAUUAGUUAGUUU-3’ |
| si3-circ-JA760602 | 5’-AAUUGAAAACAAAAUACUCAA-3’ |
| si-NC | 5’-GAGAGAUCCCAGCGCGCAGAA-3’ |
| si1-EGR1 | 5’-UUUUGUCUGCUUUCUUGUCCU-3’ |
| si-NC | 5’-GGGACUUUGCAGGCAGCGGCG-3’ |
| si1-E2F1 | 5’-UUAAAUGUUUCCAAACAGGCU-3’ |
| si-NC | 5’-CCUUUCUACGCUGGGCCGGUU-3’ |
| si1-BCL2 | 5'-UUUAUUAGUUCAACUUUCCCG-3’ |
| si2-BCL2 | 5'UAUUUUUGGCAAAAACAGGUA-3’ |
| si3-BCL2 | 5'-UUAUUUUUGGCAAAAACAGGU-3’ |
Western blot
AC16 cells were lysed by RIPA lysis buffer. Total protein was collected and then separated by 10-12% SDS-PAGE. Afterward, proteins were moved to PVDF membranes, and then the membranes were blocked with 5% skim milk. Subsequently, the membranes were incubated with primary antibodies, including EGR1 (1/1000, ab300449, Abcam, Cambridge, MA, USA), E2F1 (1/1000, ab288369, Abcam), Histone H3 (1/1000, ab1791, Abcam), GAPDH (1/1000, ab8245, Abcam) at 4°C overnight. After washing, the secondary antibody (1/2000, ab7063) were added for another two hours’ incubation. The protein levels were detected using the enhanced chemiluminescence (ECL). GAPDH was utilized as the internal reference.
Fluorescence in situ hybridization (FISH) assay
FISH assay was conducted to determine the localization of circ-JA760602 in AC16 cells. FISH probe specifically targeting circ-JA760602 were designed and synthesized (probe sequence: ACCGCTACACATGGCACATG). After AC16 cells were fixed with 4% paraformaldehyde (PFA), the obtained FISH probes were incubated with fixed cells in the hybridization buffer. DAPI was used to stain cell nuclei. Confocal laser microscopy was applied to capture fluorescent signals.
Subcellular fractionation assay
Cytoplasmic and Nuclear RNA Purification Kit (Norgenbiotek, Thorold, ON, Canada) was used for this experiment. After the cell lysates were centrifuged, cytoplasmic and nuclear parts of AC16 cells were separated. Then, RNA levels of circ-JA760602, β-actin (cytoplasmic reference) and U6 (nuclear reference) in different cellular parts were detected through qRT-PCR assay. The protein levels of EGR1 and E2F1 in cellular parts were examined through western blot by using Histone H3 as the nuclear reference and GAPDH as the cytoplasmic reference.
Immunofluorescence (IF) assay
AC16 cells were washed with PBS and fixed with 4% PFA (Sangon Biotech, Shanghai, China) fixation for 15 min at room temperature. Then, cells were treated with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) for permeabilization. After being washed with PBS, cells were incubated with Anti-EGR1 (1/100, ab300449, Abcam) and Anti-E2F1 (1/100, ab288369, Abcam) at 4°C overnight. Subsequently, the fluorescent secondary antibody was added for further incubation. The distribution of target proteins was observed under a fluorescence microscope.
Cell counting kit-8 (CCK-8) assay
CCK-8 kit was applied to measure viability of AC16 cells. In brief, cells were seeded in 96-well plates, and 10 μl of CCK-8 reagent (MedChemExpress, Monmouth Junction, NJ, USA) was added into each well at the indicated time points (0, 24, 48, 72 h). Eventually, the value of optical density (OD) was detected at 450 nm wavelength using a microplate reader (Molecular Devices, Sunnyvale, USA).
Terminal-deoxynucleotidyl transferase mediated nick end labeling (TUNEL) assay
AC16 cells were incubated in 24-well plates. Cells were fixed with 4% PFA and permeabilized with 0.5% Triton X-100. After that, cells were treated with 50 μl TdT reaction mix (Roche, Basel, Switzerland). Cell nuclei were counterstained with DAPI (Sigma-Aldrich). The images of TUNEL positive cells were captured under a fluorescence microscope.
Flow cytometry analysis
This assay was carried out based on previous description (Li et al., 2020b). Briefly, transfected AC16 cells were washed with PBS. Subsequently, Annexin V-FITC and PI were added to treat the transfected cells for 15 min in the dark. Cell apoptotic condition was analyzed using a flow cytometer.
Caspase-3 activity assay
Caspase-3 Activity Assay Kit was used in this assay according to manufacturer’s instructions. The total protein was first extracted from AC16 cells using lysis buffer. Then, protein extracts were incubated with reaction buffer and caspase-3 substrate. Finally, caspase-3 activity was detected at 405 nm wavelength with a microplate reader.
Luciferase reporter assay
To evaluate the role of circ-JA760602 in the activity of BCL2 3’UTR, the pmirGLO vector containing the sequence of BCL2 3’UTR was constructed. Similarly, the pGL3 vector (E1751, Promega, Madison, WI, USA) inserted with the sequence of BCL2 promoter and the pmirGLO vector (E1330, Promega) inserted with the sequence of BCL2 were constructed. The synthesized plasmid (pmirGLO-BCL2 3’UTR or pGL3-BCL2-promoter) was co-transfected with si-NC/si1-circ-JA760602 into hypoxia-induced AC16 cells. Additionally, pGL3-BCL2-promoter was also co-transfected with si1-EGR1/si1-E2F1 using Lipofectamine 2000 (Invitrogen) to detect the influence of EGR1/E2F1 knockdown on BCL2 transcription. Following 48 hours’ transfection, Luciferase Reporter Gene Assay kit (E1910, Promega) was used to examine the luciferase activities, and the Rnilla luciferase activity was used as negative control.
Chromatin immunoprecipitation (ChIP) assay
This assay was conducted to assess the combinations between EGR1/E2F1 and BCL2 promoter. The crosslinked chromatin was cut into fragments (200-1000 bp). Subsequently, primary antibodies, including Anti-EGR1 (1/200, ab300449, Abcam), Anti-E2F1 (1/200, Millipore, Bedford, MA, USA), and control Anti-IgG (1/100, Millipore) were incubated with the obtained cell lysates, respectively. After magnetic beads were added to precipitate DNA-protein complex, the purified DNA enriched in the complex was isolated and quantified through qRT-PCR.
RNA binding protein immunoprecipitation (RIP)
Cells were lysed by RIP lysis buffer. The obtained cell lysates were incubated with specific primary antibodies, including Anti-EGR1 (1/100, ab300449, Abcam), Anti-E2F1 (1/100, Millipore), and control Anti-IgG (1/100, Millipore), and then with magnetic beads at 4°C overnight. Following purification, the enrichment of RNAs precipitated with beads was examined through qRT-PCR.
RNA pull-down assay
Biotinylated (Bio)-NC and Bio-circ-Ja760602 were generated in advance. Lysed AC16 cells were incubated with the synthesized biotin-labeled probes and streptavidin magnetic beads. After purification, the proteins pulled down by beads were detected through western blot.
Statistical analysis
Each experiment was conducted in triplicate. The data were analyzed using SPSS software, and the results were displayed in the form of mean ± standard deviation (SD). Student’s t-test was applied to compare statistic difference between two groups. One-way analysis of variance (ANOVA) was utilized to analyze differences among multiple groups. Statistical significance of data was defined as P value less than 0.05.
Declarations
Funding
This work was supported by Hefei Hospital Affiliated to Medical University of Anhui (No. 2019xkj086).
Abbreviations
ActD, Actinomycin D ; AMI, Acute myocardial infarction ; ANOVA, Analysis of variance ; ATCC, American Type Culture Collection ; CCK-8, Cell counting kit-8 ; ChIP, Chromatin immunoprecipitation ; circRNA, Circular RNA ; CVD, Cardiovascular disease ; DMEM, Dulbecco’s Modified Eagle’s Medium ; ECL, Enhanced chemiluminescence ; FBS, Fetal bovine serum ; FISH, Fluorescence in situ hybridization ; IF, Immunofluorescence ; NC, Negative control ; OD, Optical density ; PFA, Paraformaldehyde ; qRT-PCR, Quantitative real-time polymerase chain reaction ; RBP, RNA binding protein ; RIP, RNA binding protein immunoprecipitation ; SD, Standard deviation ; siRNA, Small interfering RNA ; TUNEL, Terminal-deoxynucleotidyl transferase mediated nick end labeling ;References
Bhaskar Rao D., Panneerpandian P., Balakrishnan K., Ganesan K. (2021). YY1 regulated transcription‐based stratification of gastric tumors and identification of potential therapeutic candidates. Journal of Cell Communication and Signaling 15: 251-267.
Cai X., Li B., Wang Y., Zhu H., Zhang P., Jiang P., Yang X., Sun J., Hong L., Shao L. (2021). CircJARID2 Regulates Hypoxia-Induced Injury in H9c2 Cells by Affecting miR-9-5p–Mediated BNIP3. Journal of Cardiovascular Pharmacology 78: e77-e85.
Cao S., Li C., Li L., Zhou G., Jiang Y., Feng J. (2022b). Circular RNA hsa_circ_0000848 Regulates Cardiomyocyte Proliferation and Apoptosis Under Hypoxia via Recruiting ELAVL1 and Stabilizing SMAD7 mRNA. The Anatolian Journal of Cardiology 26: 189-197.
Cao T., Lu Y., Wang Q., Qin H., Li H., Guo H., Ge M., Glass S. E., Singh B., Zhang W., Dong J., Du F., Qian A., Tian Y., Wang X., Li C., Wu K., Fan D., Nie Y., Coffey R. J., Zhao X. (2022a). A CGA/EGFR/GATA2 positive feedback circuit confers chemoresistance in gastric cancer. Journal of Clinical Investigation 132: e154074.
Chen C., Yuan W., Zhou Q., Shao B., Guo Y., Wang W., Yang S., Guo Y., Zhao L., Dang Q., Yang X., Wang G., Kang Q., Ji Z., Liu J., Sun Z. (2021b). N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics 11: 4298-4315.
Chen T., Zhang N., Wang H., Hu S., Geng X. (2021). Knockdown of circROBO2 attenuates acute myocardial infarction through regulating the miR-1184/TRADD axis. Molecular Medicine 27: 21.
Chen Y., Miao J., Lou G. (2021a). Knockdown of circ-FURIN suppresses the proliferation and induces apoptosis of granular cells in polycystic ovary syndrome via miR-195-5p/BCL2 axis. Journal of Ovarian Research 14: 156.
Feng J., Zhan J., Ma S. (2021). LRG1 promotes hypoxia-induced cardiomyocyte apoptosis and autophagy by regulating hypoxia-inducible factor-1α. Bioengineered 12: 8897-8907.
Hong X., Li Q., Li J., Chen K., He Q., Zhao Y., Liang Y., Zhao Y., Qiao H., Liu N., Ma J., Li Y. (2022). CircIPO7 Promotes Nasopharyngeal Carcinoma Metastasis and Cisplatin Chemoresistance by Facilitating YBX1 Nuclear Localization. Clinical Cancer Research 28: 4521-4535.
Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W., Liao Y., Bin J. (2019). Loss of Super-Enhancer-Regulated circRNA Nfix Induces Cardiac Regeneration After Myocardial Infarction in Adult Mice. Circulation 139: 2857-2876.
Jiao L., Li M., Shao Y., Zhang Y., Gong M., Yang X., Wang Y., Tan Z., Sun L., Xuan L., Yu Q., Li Y., Gao Y., Liu H., Xu H., Li X., Zhang Y., Zhang Y. (2019). lncRNA-ZFAS1 induces mitochondria-mediated apoptosis by causing cytosolic Ca2+ overload in myocardial infarction mice model. Cell Death & Disease 10: 942.
Kapur N. K., Thayer K. L., Zweck E. (2020). Cardiogenic Shock in the Setting of Acute Myocardial Infarction. Methodist DeBakey Cardiovascular Journal 16: 16.
Kloner R. A., Dai W., Hale S. L., Shi J. (2016). Approaches to Improving Cardiac Structure and Function During and After an Acute Myocardial Infarction. Journal of Cardiovascular Pharmacology and Therapeutics 21: 363-367.
Lafita-Navarro M. C., Liaño-Pons J., Quintanilla A., Varela I., Blanco R., Ourique F., Bretones G., Aresti J., Molina E., Carroll P., Hurlin P., Romero O. A., Sanchez-Céspedes M., Eisenman R. N., Delgado M. D., León J. (2020). The MNT transcription factor autoregulates its expression and supports proliferation in MYC-associated factor X (MAX)-deficient cells. Journal of Biological Chemistry 295: 2001-2017.
Li DJ, Wang X, Yin WH, Niu K, Zhu W, Fang N, (2020b). MiR-199a-5p suppresses proliferation and invasion of human laryngeal cancer cells.. European review for medical and pharmacological sciences 24: 12200-12207.
Li Y., Ren S., Xia J., Wei Y., Xi Y. (2020a). EIF4A3-Induced circ-BNIP3 Aggravated Hypoxia-Induced Injury of H9c2 Cells by Targeting miR-27a-3p/BNIP3. Molecular Therapy - Nucleic Acids 19: 533-545.
Liu C.X., Chen L.L. (2022). Circular RNAs: Characterization, cellular roles, and applications. Cell 185: 2016-2034.
Liu H.T., Liu S., Liu L., Ma R.R., Gao P. (2018). EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer. Cancer Research 78: 5877-5890.
Lu X., Gao H., Zhu B., Lin G. (2021). Circular RNA circ_RANBP9 exacerbates polycystic ovary syndrome via microRNA-136-5p/ XIAP axis . Bioengineered 12: 6748-6758.
Luo D., Ge W. (2020). MeCP2 Promotes Colorectal Cancer Metastasis by Modulating ZEB1 Transcription. Cancers 12: 758.
Ma J., He Z., Zhang H., zhang W., Gao S., Ni X. (2021). SEC61G promotes breast cancer development and metastasis via modulating glycolysis and is transcriptionally regulated by E2F1. Cell Death & Disease 12: 550.
Peng W.X., He R.Z., Zhang Z., Yang L., Mo Y.Y. (2019). LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription. Oncogene 38: 6770-6780.
Qi Y., Mo K., Zhang T. (2021). A transcription factor that promotes proliferation, migration, invasion, and epithelial–mesenchymal transition of ovarian cancer cells and its possible mechanisms. BioMedical Engineering OnLine 20: 83.
Tang D., Li Z.Y., Gijsen F., Giddens D. P. (2015). Cardiovascular diseases and vulnerable plaques: data, modeling, predictions and clinical applications. BioMedical Engineering OnLine 14: S1.
Wang S., Cheng Z., Chen X., Lu G., Zhu X., Xu G., (2021a). CircUBXN7 mitigates H/R-induced cell apoptosis and inflammatory response through the miR-622-MCL1 axis. American journal of translational research 13: 8711-8727.
Wang S., Li L., Deng W., Jiang M. (2021b). CircRNA MFACR Is Upregulated in Myocardial Infarction and Downregulates miR-125b to Promote Cardiomyocyte Apoptosis Induced by Hypoxia. Journal of Cardiovascular Pharmacology 78: 802-808.
Wu T., Wu D., Wu Q., Zou B., Huang X., Cheng X., Wu Y., Hong K., Li P., Yang R., Li Y., Cheng Y. (2017). Knockdown of Long Non‐Coding RNA‐ZFAS1 Protects Cardiomyocytes Against Acute Myocardial Infarction Via Anti‐Apoptosis by Regulating miR‐150/CRP. Journal of Cellular Biochemistry 118: 3281-3289.
Yan X., Hou J. (2021). miR-22 Host Gene Enhances Nuclear Factor-kappa B Activation to Aggravate Hypoxia-induced Injury in AC16 Cardiomyocytes. Cell Transplantation 30: 096368972199032.
Yang J., He W., Gu L., Long J., Zhu L., Zhang R., Zhao Z., Xu B., Nan A., Su L. (2022). CircUSP36 attenuates ischemic stroke injury through the miR-139-3p/SMAD3/Bcl2 signal axis. Clinical Science 136: 953-971.
Yu F., Shi G., Cheng S., Chen J., Wu S.Y., Wang Z., Xia N., Zhai Y., Wang Z., Peng Y., Wang D., Du J. X., Liao L., Duan S.Z., Shi T., Cheng J., Chiang C.M., Li J., Wong J. (2018). SUMO suppresses and MYC amplifies transcription globally by regulating CDK9 sumoylation. Cell Research 28: 670-685.
Zang J., Lu D., Xu A. (2020). The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. Journal of Neuroscience Research 98: 87-97.
Zhang Y., Huang Y.X., Wang D.L., Yang B., Yan H.Y., Lin L.H., Li Y., Chen J., Xie L.M., Huang Y.S., Liao J.Y., Hu K.S., He J.H., Saw P. E., Xu X., Yin D. (2020). LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics 10: 10823-10837.
Zhao B., Li G., Peng J., Ren L., Lei L., Ye H., Wang Z., Zhao S. (2021). CircMACF1 Attenuates Acute Myocardial Infarction Through miR-500b-5p-EMP1 Axis. Journal of Cardiovascular Translational Research 14: 161-172.