Int. J. Dev. Biol. 68: 85 - 91 (2024)
Expression analysis of thg1l during Xenopus laevis development
Open Access | Developmental Expression Pattern | Published: 15 July 2024
Abstract
The tRNA-histidine guanylyltransferase 1-like (THG1L), also known as induced in high glucose-1 (IHG-1), encodes for an essential mitochondria-associated protein highly conserved throughout evolution, that catalyses the 3′–5′ addition of a guanine to the 5′-end of tRNA-histidine (tRNAHis). Previous data indicated that THG1L plays a crucial role in the regulation of mitochondrial biogenesis and dynamics, in ATP production, and is critically involved in the modulation of apoptosis, cell-cycle progression and survival, as well as in cellular stress responses and redox homeostasis. Dysregulations of THG1L expression play a central role in various pathologies, including nephropathies, and neurodevelopmental disorders often characterized by developmental delay and cerebellar ataxia. Despite the essential role of THG1L, little is known about its expression during vertebrate development. Herein, we examined the detailed spatio-temporal expression of this gene in the developing Xenopus laevis. Our results show that thg1l is maternally inherited and its temporal expression suggests a role during the earliest stages of embryogenesis. Spatially, thg1l mRNA localizes in the ectoderm and marginal zone mesoderm during early stages of development. Then, at tadpole stages, thg1l transcripts mostly localise in neural crests and their derivatives, somites, developing kidney and central nervous system, therefore largely coinciding with territories displaying intense energy metabolism during organogenesis in Xenopus.
Keywords
Post-transcriptional modifications, tRNA, mitochondria, Spinocerebellar Ataxia
Introduction
Post-transcriptional modifications of tRNAs are highly evolutionary conserved mechanisms required for accurate translation. These modifications play a number of different roles, such as regulating the decay of both pre-tRNA and mature tRNA, controlling specificity and accuracy of mRNA decoding, and modulating the efficiency and specificity of tRNA aminoacylation (Phizicky and Hopper, 2023). Several data have shown that impairments of tRNA post-transcriptional modification machinery have a central role in various pathological conditions, such as neurological disorders and cancer, and also affect embryonic development (Balke et al., 2015; Blaze and Akbarian, 2022). The tRNA-histidine guanylyltransferase 1-like (THG1L), also known as “induced in high glucose-1” (IHG-1), encodes for a highly evolutionarily conserved essential protein associated with the inner mitochondrial membrane (Bhreathnach et al., 2017; Hickey et al., 2011) that catalyses the 3′–5′ addition of a guanine to the 5′-end of tRNA-histidine (tRNAHis) (Murphy et al., 2013). With few exceptions, the unusual 5′-end consisting of an extra guanylate residue called G–1 in the acceptor stem is required for tRNAHis aminoacylation by histidyl-tRNA synthetases, both in prokaryotes and in eukaryotes (Rosen et al., 2006). Although its essential role has been underlined, THG1L is still a little-studied gene, whose molecular pathways and involvement in physiological and pathological conditions are far from being elucidated.
To date, previous studies have described THG1L as a key regulator of mitochondrial function that, by stabilizing the transcriptional cofactor peroxisome proliferator–activated receptor-γ coactivator 1α (PGC-1α), increases the mitochondrial respiratory capacity, ATP production, biogenesis and fusion, therefore influencing cell survival in conditions of oxidative stress (Edvardson et al., 2016; Hickey et al., 2011; Hickey et al., 2014). Moreover, in vitro experiments on HeLa cells have suggested that IHG-1 may have a role in the regulation of cell cycle progression, cell morphology, and proliferation (Guo et al., 2004). In the same line, in silico analysis have reported that THG1L functionally interacts with various key mediators involved in apoptosis, proliferation, and also with regulators of cellular stress response and redox homeostasis (Bhreathnach et al., 2017). Previous works have indicated that THG1L expression dysregulations have a central role in the pathogenesis of tubulointerstitial fibrosis promoted by the major pro-fibrotic key mediator, the transforming growth factor 1-β (TGF1-β), in both diabetic nephropathy in humans and in experimental renal fibrosis (Bhreathnach et al., 2017; Corcoran et al., 2013; Murphy et al., 2008). Although far to be elucidated, it has been suggested that THG1L plays a critical role during development. In particular, specific THG1L variants have been identified in patients showing various degrees of developmental delay and cerebellar ataxia, sometimes associated with microcephaly, cerebellar hypoplasia, dysarthria, pyramidal signs and epileptic encephalopathy, as well as, in rare cases, other multisystem abnormal manifestations (Edvardson et al., 2016; Han et al., 2023; Rabin et al., 2021; Shaheen et al., 2019; Walker et al., 2019). Indications of a potential involvement of thg1l in central nervous system (CNS) development have been also provided by microarray analyses in developing Xenopus laevis embryos, showing that thg1l is among the genes coherently regulated by the paired-like homeodomain transcription factor Rax/Rx1 (Giudetti et al., 2014), a well-known critical factor for vertebrate retina specification and morphogenesis, which also plays a key role in brain development (Terada et al., 2006). Finally, considering that dysregulations of mitochondrial function have an active role in the pathogenesis of several diseases, including neurodegenerative, cardiac, skeletal muscle disorders (Chen et al., 2023; Schulz and Schluter, 2023), as well as in embryo development (Bruna de Lima et al., 2023; May-Panloup et al., 2021), clearly emerges the importance of research aimed at increasing knowledge about this gene. To our knowledge, the expression pattern of THG1L during embryogenesis is a topic that has been poorly investigated. As Xenopus laevis is a well-known experimental model in biomedical research and development, we have performed a detailed spatio-temporal expression analysis of thg1l on developing Xenopus, with the aim to increase the knowledge on this tRNA post-transcriptional modifier and provide a foundation for further studies on its functional role.
Results and Discussion
Temporal expression of thg1l
To analyze the temporal expression pattern of thg1l during embryonic development, we performed real-time reverse transcription-polymerase chain reaction (RT-qPCR) assay on cDNAs obtained from Xenopus embryos at different developmental stages. Our results show that this gene is expressed during embryonic life, in line with some evidence provided by in situ hybridization (ISH) in zebrafish (Thisse and Thisse, 2004), as well as by microarray in Xenopus laevis (Giudetti et al., 2014). In detail, our temporal expression analysis (Fig. 1) shows that thg1l transcripts are already detectable at blastula 64-cell stage (stage, St. 7), and therefore before mid-blastula transition (St. 8 according to Newport and Kirschner (1982)), indicating the presence of maternal transcripts, as also suggested by previous transcriptomic data (Session et al., 2016; Yanai et al., 2011), and a potential role for thg1l during the earliest phases of embryonic development. In the same line, maternal expression of thg1l has been reported in zebrafish (Thisse and Thisse, 2004). Maternal transcripts of other highly conserved tRNA post-transcriptional modifiers have been reported in Xenopus and zebrafish as, for instance, GTP-binding protein 3 (gtpbp3) (Chen et al., 2016), tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (trmu) (Zhang et al., 2018), tRNA nucleotidyl transferase 1 (trnt1) (DeLuca et al., 2016) and tRNA (guanine(37)-N1)-methyltransferase 5 (trmt5) (White et al., 2017). Interestingly, in line with data on THG1L, defects in these genes are responsible for rare syndromes characterized by severe pathological traits in humans, such as varying degrees of brain defects, developmental delay, intellectual disability, seizure, dysarthria and ataxia (Argente-Escrig et al., 2022; Chakraborty et al., 2014; Reinhart et al., 1993; Sasarman et al., 2015; Wiseman et al., 2013; Zhang et al., 2023). Successively, thg1l expression levels in Xenopus laevis are quickly reduced, probably due to the consumption of maternal mRNA, reaching the lowest expression level by gastrula stage (St. 11), and then remaining almost unchanged until St. 42 tadpole. A similar temporal expression profile was previously reported, by RNA-sequencing analysis, in the Xenbase database for thg1l (Fisher et al., 2023) and also for other tRNA modifiers such as trnt1, trmu, and trmt5 (Session et al., 2016; Yanai et al., 2011), indicating that specific post-transcriptional processing of tRNAs, including the 3′–5′ addition of a guanine to the 5′-end of tRNA-histidine (tRNAHis), is regulated in a time-dependent manner, and may have a fundamental role especially in the initial phases of vertebrate embryogenesis. As THG1L influences mitochondrial activity (Edvardson et al., 2016; Hickey et al., 2011; Hickey et al., 2014), this possibility is in line with the fact that the function of mitochondria is finely modulated during development, playing a pivotal role during the earliest stages, as previously reported in various experimental models, including Xenopus laevis (Han et al., 2018; Klymkowsky, 2011).
Fig. 1. RT-qPCR analysis of thg1l expression during development.
cDNAs were synthesized from embryos and larvae at different developmental stages (St., indicated at the bottom of the panel) and amplified using specific primers for thg1l, as well as sub1.S, slc35b1.L and ppp1ca.L, used for normalization. As technical control, reverse transcriptase was omitted. Asterisks (*) indicate statistically significant difference between each stage and St. 7. Data are expressed as mean ± standard error of the mean; n, number of samples analysed. ANOVA followed by Tukey post-hoc test: ****p < 0.0001.
Spatial expression of thg1l
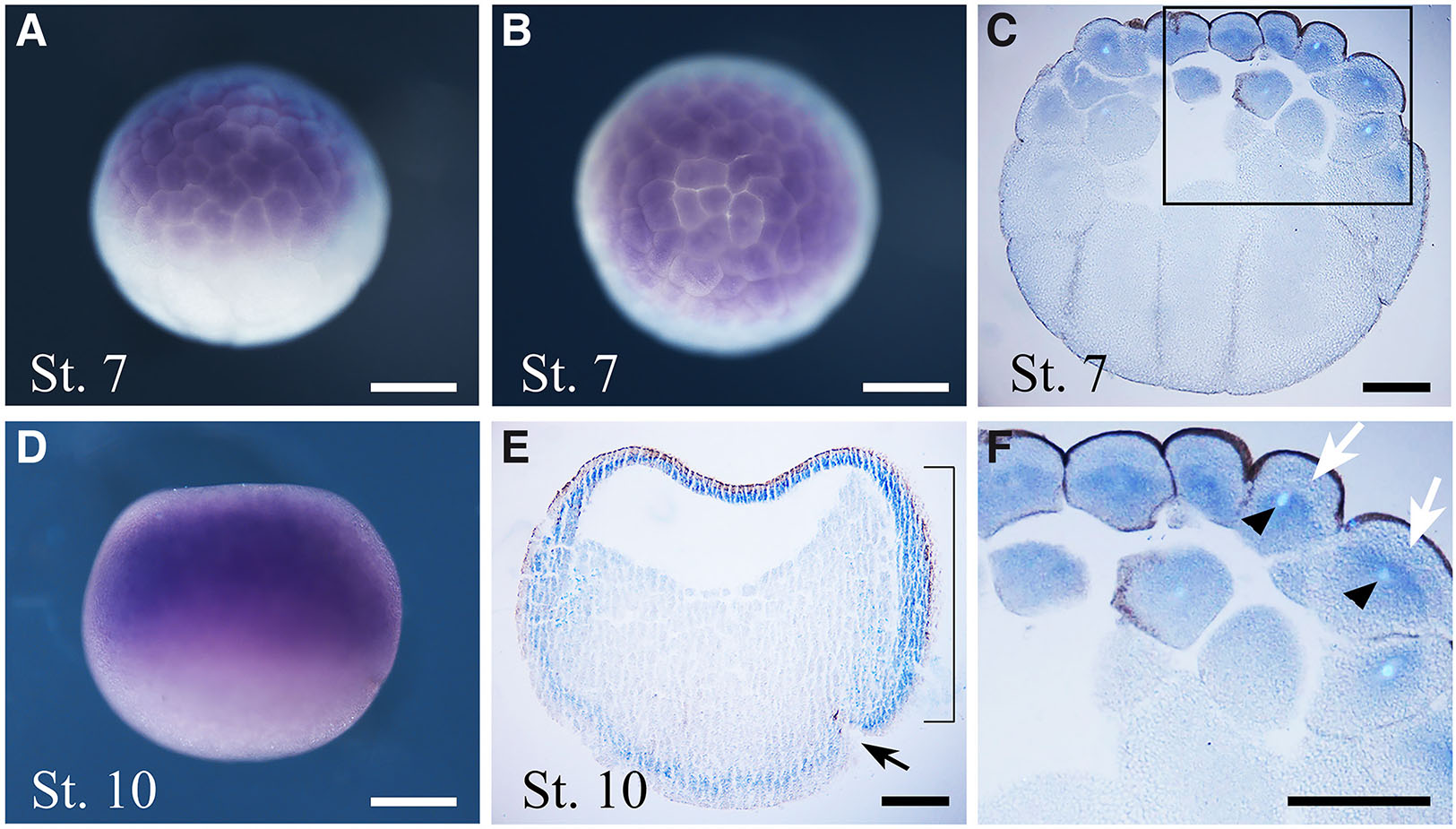
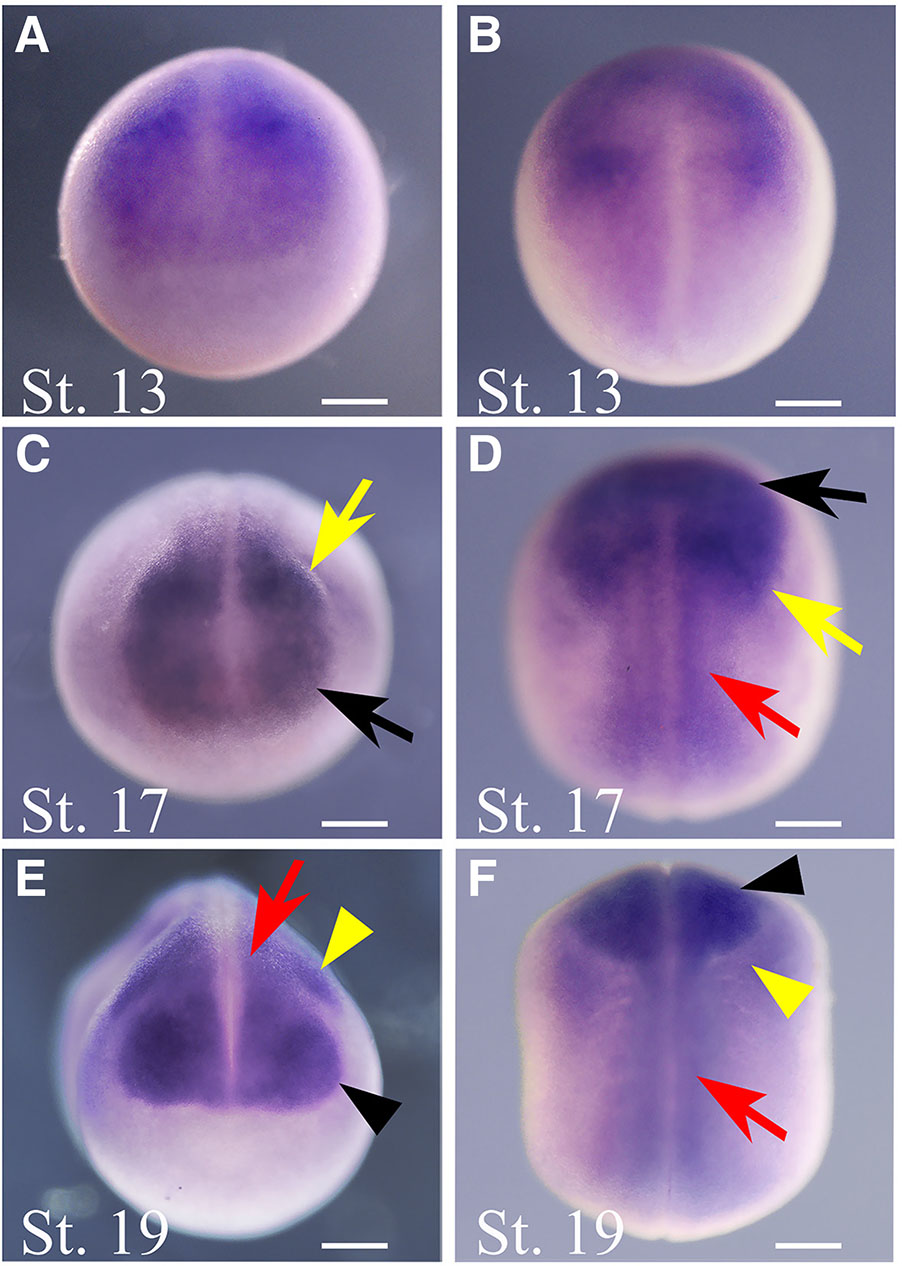
To determine the spatial expression of thg1l we performed ISH throughout development. During the earliest blastula stages (St. 7), before mid-blastula transition, maternal thg1l transcripts localise in the animal hemisphere (Fig. 2 A,B,C,F). In particular, the hybridization signal appears mostly localised in the basal portion of animal blastomeres at this developmental stage (Fig. 2F) suggesting the possibility that the asymmetric localization of thg1l mRNA at early developmental stages may be involved in cell-fate determination as reported for various mRNA asymmetrically distributed in the cells (Skamagki et al., 2013). At early gastrula St. 10 (Fig. 2 D,E), thg1l is expressed in the ectoderm and in the presumptive mesoderm of the marginal zone. At early neurula stage (St.13, Fig. 3A,B), thg1l signal localizes in anterior regions of the neural plate and, although at significantly lower level, also in the posterior neural plate. This pattern becomes more demarcated at mid-neurula (St. 17, Fig. 3 C,D) when the expression of thg1l increases in the most anterior region of the neural plate and in territories of neural crests, while additional lower signal continues to be detectable in the in the posterior neural plate. Successively, after neural tube closure at late-neurula (St. 19, Fig. 3 E,F), thg1l hybridization signal is observed in the neural tube, in migrating neural crest cells, and in optic vesicles.
Fig. 2. In situ hybridisation analysis of thg1l during segmentation and gastrulation.
Embryo developmental stages (St.) are indicated at the bottom left corner of each panel. (A,D) Lateral view, animal pole to the top and vegetal pole to the bottom, and (B) animal view, of whole-mount stained embryos. (C,E) Sagittal sections from whole-mount embryos, animal pole to the top and vegetal pole to the bottom. Black arrow in (E) points to the dorsal lip of the blastopore. (F) Magnified view of the boxed region indicated in (C), showing the intracellular localization of thg1l transcripts in animal blastomeres. White arrows show absence of the transcript in the apical portion of cytoplasm, whereas black arrowheads indicate Hoechst-stained nuclei. Bracket in (E) indicate the dorsal involuting mesoderm. Scale bars: 200 µm in (A,B,D); 150 µm in (C,E,F).
Fig. 3. In situ hybridisation analysis of thg1l during neurulation.
Embryo developmental stages (St.) are indicated at the bottom left corner of each panel. (A,C,E) Frontal view, dorsal to the top, and (B,D,F) dorsal view, frontal to the top, of whole-mount hybridized embryos. Black arrows indicate the anterior neural plate. Yellow arrows point to the neural crests. Red arrows indicate the neural tube. Black arrowheads point to the optic vesicles. Yellow arrowheads indicate migrating neural crest cells. Scale bar: 250 µm.
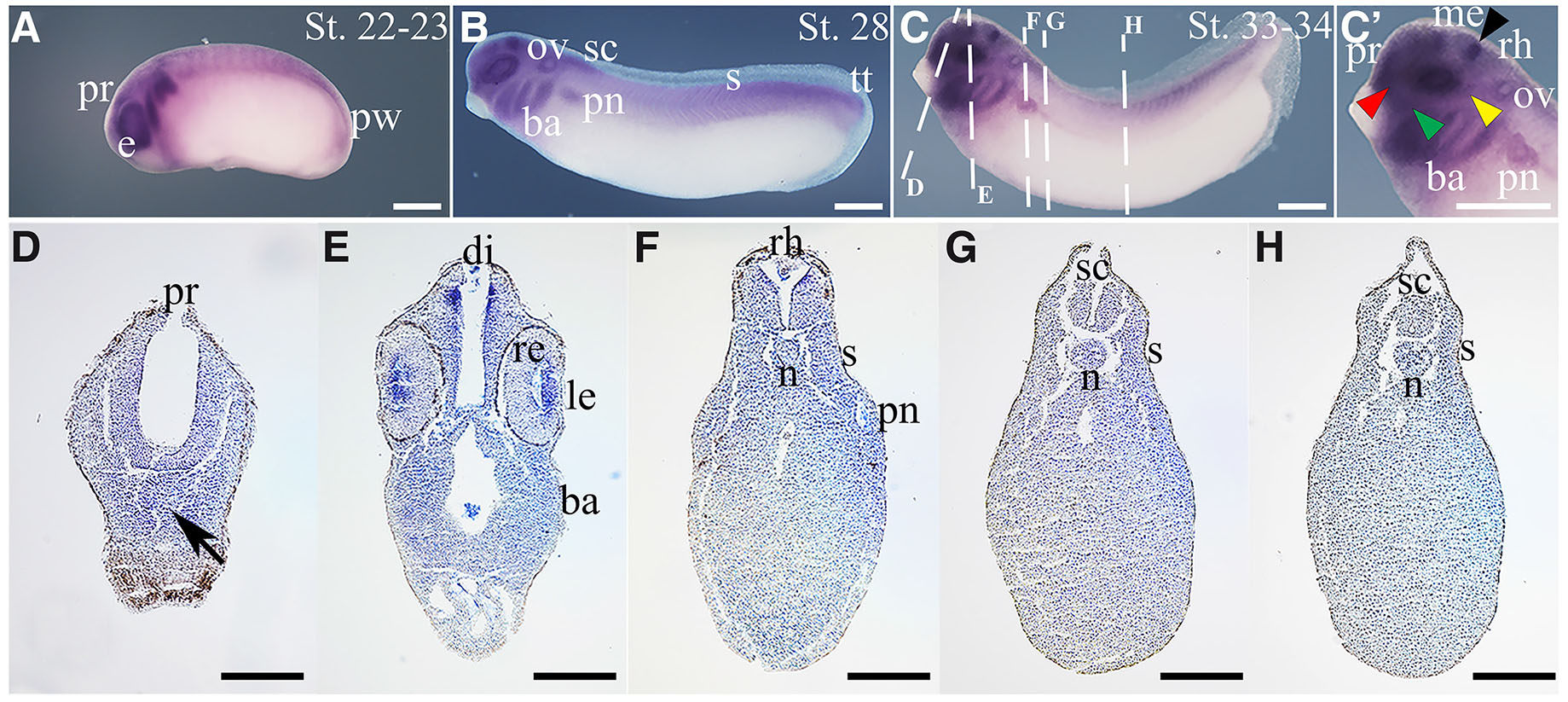
At early tailbud (St. 22-23, Fig. 4A), thg1l transcripts mostly localise in eye vesicles and in migrating neural crest material, with a weak signal also detectable in the prosencephalon; therefore, all structures originating from the territories where thg1l is expressed at neurula stage. Additionally, a weak expression is present in somites and in the posterior region of the trunk, including the posterior wall.
Fig. 4. In situ hybridisation analysis of thg1l in tailbud embryos.
Embryo developmental stages (St.) are indicated at the top right corner of each panel. (A,B,C) Lateral views of whole-mount hybridized embryos, anterior to the left; (C') magnified view of (C). Dashed lines in (C) indicate the section planes shown in (D-H). (D-H) Transverse sections from whole-mount St. 33-34 embryos, dorsal to the top. Black arrow in (D) points to the Rathke’s pouch. Black arrowhead in (C') indicates the midbrain-hindbrain boundary; yellow arrowhead indicates lateral line placode; green arrowhead indicates epibranchial placode, and red arrowhead indicates olfactory placode. Scale bars: 500 µm in (A-C, C’); 250 µm in (D-H). ba, branchial arches; di, diencephalon; e, eye; le, lens; me, mesencephalon; n, notochord; ov, otic vesicle; pn, pronephros; pr, prosencephalon; pw, posterior wall; re, retina; rh, rhombencephalon; s, somites; sc, spinal cord; tt, tail tip.
By St. 28 (Fig. 4B), thg1l is clearly recognizable in the prosencephalon, rostral region of the spinal cord, branchial arches, developing eye, otic vesicles, pronephros, somites in the posterior half of the trunk, and in the growing tail. As shown in Fig. 4C, a similar expression pattern is mostly maintained at later tailbud stage (St.33/34), when a hybridization signal becomes observable in the midbrain-hindbrain boundary, and a weak signal is also distinguishable in cranial placodes, including olfactory, epibranchial and lateral line placode, mesencephalon and rhombencephalon. Additionally, histological sections at this stage (Fig. 4 D-H) show a clear signal in lens, whereas a faint signal is observed in the retina, Rathke’s pouch and posterior region of the notochord, suggesting that thg1l expression level in these regions is likely to be weak. Suggestive of a possible influence of thg1l on mitochondrial activity also during embryogenesis, a previous work in Xenopus laevis reported that the nervous system, eye, skeletal muscle and kidney tissues (territories in which thg1l is clearly expressed), are sites with high energy requirements during organogenesis. In addition, the same authors have shown that the spatial expression pattern of GRIM-19, a newly identified subunit of the Mitochondrial Respiratory Chain (MRC) complex I, which plays crucial roles in basic biological processes through the production of cellular energy, generation of reactive oxygen species, and initiation of apoptosis, is largely similar to the one we report for thg1l (Chen et al., 2007).
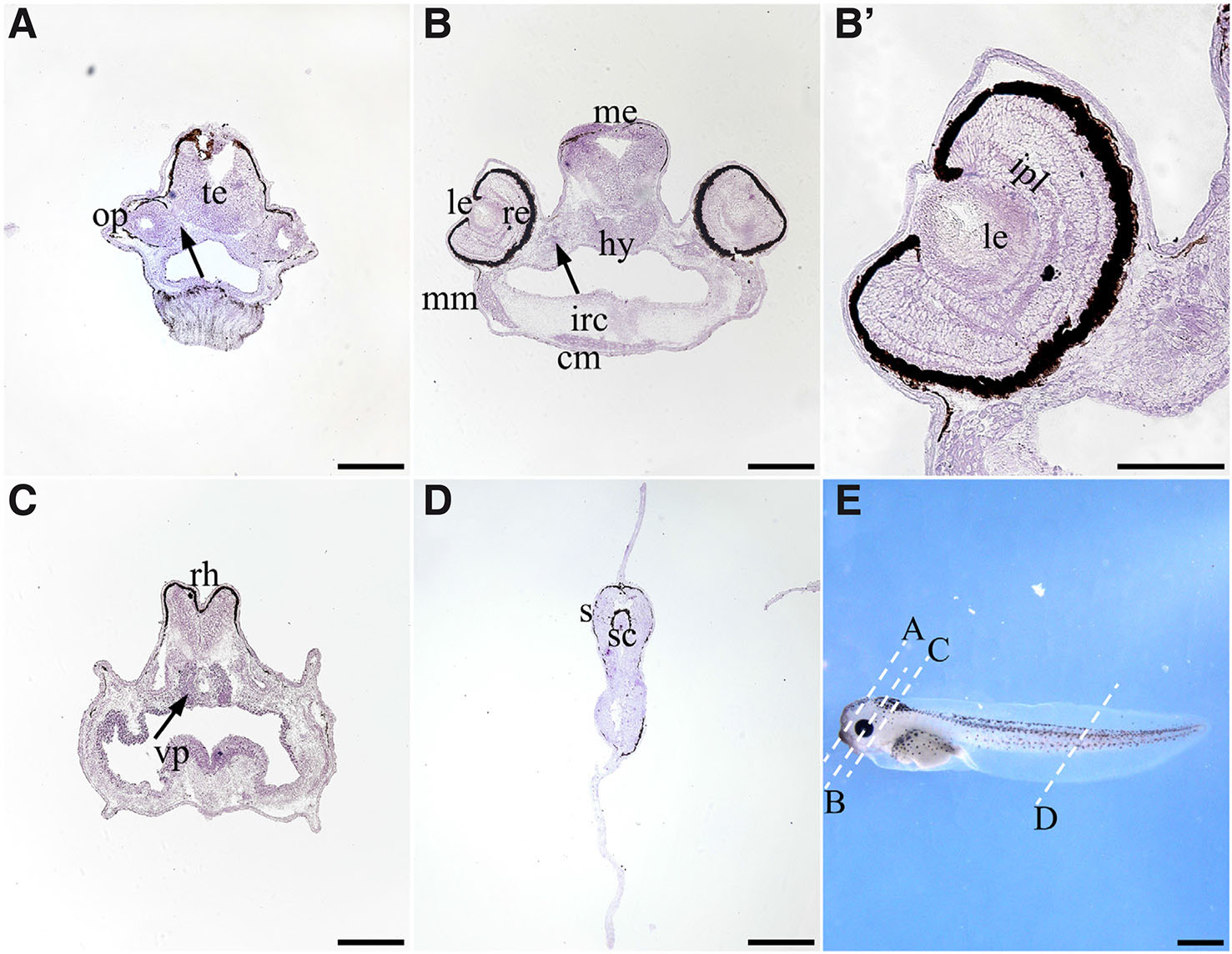
As shown in (Fig. 5), in the head region of St. 42 tadpoles, thg1l transcripts localise in the olfactory pits and Jacobson's organ, telencephalon, hypothalamus, mesencephalon and rhombencephalon (Fig. 5 A-C). In addition, the hybridization signal is clearly observed in the velar plate, parachordal cartilages, cranial muscles including mandibular muscles, as well as a weak expression is localized in the infrarostral cartilage (Fig. 5 B,C). Moreover, in the eye region, at the time when the retina is mature, thg1l transcripts mostly localize in the lens, inner plexiform layer and external ocular muscles, although a weak signal is diffused in all retinal layers (Fig. 5 B,B’). Finally, in the trunk-caudal region, thg1l expression was observed in the spinal cord and somites (Fig. 5D).
Fig. 5. In situ hybridisation analysis of thg1l in stage 42 larvae.
(A,B,B’,C,D) Transverse cryosections, dorsal to the top. (E) Lateral views of the larvae, anterior to the left. Dashed lines in (E) indicate the section planes shown in (A,B,B′,C,D). Black arrows indicate the Jacobson's organ in (A), external ocular muscles in (B), and parachordal cartilages in (C); (B′) high magnification of the eye represented in (B). Scale bars, 200 μm in (A,B,C,D); 1 mm in (E) and 100 μm in (B′). cm, cranial muscles; hy, hypothalamus; ipl, inner plexiform layer; irc, infrarostral cartilage; le, lens; me, mesencephalon; mm, mandibular muscles; op, olfactory pit; te, telencephalon; re, retina; rh, rhombencephalon; s, somites; sc, spinal cord; vp, velar plate.
In conclusion, our study elucidates the previously undescribed spatio-temporal expression pattern of thg1l in Xenopus laevis. The dynamic expression of the tRNA-histidine guanylyltransferase mRNA, together with the activity of its encoded protein, suggest an important role for thg1l during the development of various organs and tissues. Moreover, the differential regulation of thg1l in different cells during development is necessary for proper tissues morphogenesis and function.
Materials and Methods
Total RNA extraction and RT-qPCR analysis
Xenopus laevis embryos were generated and staged in accordance with published works (Messina et al., 2015; Newport and Kirschner, 1982). For each biological sample, total RNA was extracted from a pool of six-ten embryos, using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). Phenol-chloroform extraction was performed to isolate RNA that successively was precipitated and purified using RNAeasy Plus MiniKit (Qiagen, Venlo, The Netherlands), in line with manufacturer’s instructions. DeNovix™ (Wilmington, DE, USA) spectrophotometer was used to measure concentration and purity of total RNA, while agarose-gel electrophoresis was performed to evaluate RNA integrity. According to manufacturer’s instructions, first-strand cDNA was synthesised using QuantiTect Reverse Transcription Kit (Qiagen).
Relative measurements of mRNA expression levels were performed by RT-qPCR using the SYBR Green method (SensiMix SYBR kit, Bioline, London, UK) on a QuantStudio 3 system (ThermoFisher), following the manufacturer’s protocol. Transcript levels of thg1l were normalised on the expression of three reference genes (sub1.S, slc35b1.L and ppp1ca.L), in line with previous works (Martini et al., 2023; Mughal et al., 2018; Vandesompele et al., 2002), and calculated as 2-(ΔCT). Specific primer sequences were designed using the Primer-BLAST tool in the NCBI browser. Specificity and efficiency of primer-pairs were also evaluated before RT-qPCR analysis. Primer-pairs showing 100% efficiency were chosen. Additionally, to exclude genomic DNA contamination, reverse transcriptase obtained without reverse transcriptase was used as control. Primers sets used in this work are shown below, along with the sizes of their respective expected amplification products:
thg1L: Fw 5’- CTATCTGCCTGGCCTTTGGG -3’, Rv 5’- GTGGGTCATAAACTTGCTCGC -3’ (98 bp);
sub1.S: Fw 5’-GCAGGAGAAATGAAGCCAGG-3’, Rv 5’-CCGACATCTGCTCCTTCAGT-3’ (79 bp);
slc35b1.L: Fw 5’-CGCATTTCCAAACAGGCTCC-3’, Rv-5’-CAAGAAGTCCCAGAGCTCGC-3’ (107 bp);
ppp1ca.L: Fw 5’-ACGAGTCTCTCATGTGCTCC-3’, Rv-5’-CAGAGCTGGGAGGGGTCATT-3’ (140 bp).
In addition to the canonical thg1l gene sequence (NM_001095128.1), four predicted transcript variants are reported (XM_041585200.1, XM_018251143.2, XM_041585201.1, XM_041585202.1). thg1l forward and reverse primer anneal to exon 3 and to the junction between exon 3 and 4 of the canonical gene, respectively. Both regions bound by forward and reverse primers are conserved also in all predicted transcript variants.
RT-qPCR data were expressed as Mean ± standard error of the mean (SEM) from n=6 samples for each embryonic stage in three independent experiments. Statistical analysis was performed using the software GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, USA) and, after verification of the normal distribution, ANOVA followed by post-hoc Tukey’s Multiple comparison test was used. Differences were considered significant when p < 0.05.
In situ hybridization
thg1l antisense RNA probe was transcribed in vitro from the cDNA clone IMAGE:4970193 (GenBank Acc. n. BC085067.1), by plasmid linearization with SalI followed by T7 RNA polymerase transcription. To evaluate the strength of the hybridization signal compared to potential background staining, a thg1l sense probe was transcribed after plasmid linearization with XhoI, followed by transcription with SP6 RNA polymerase, and then assessed (Supplementary Fig. S1). Whole-mount ISH has was performed as previously described (Giannaccini et al., 2013), and BM Purple substrate (Roche, Indianapolis, IN, USA) was used for signal detection. Images of whole-mount embryos were acquired by a Nikon SMZ18 stereomicroscope (Nikon Corporation, Minato, TO, Japan) connected to a Nikon DS-Ri3 digital camera and equipped with the software NIS-Elements AR 5.11.03. In specific cases, whole mount hybridized embryos were sectioned using a microtome to obtain 20 µm-thick sections from paraffin-embedded whole-mount preparations. Sections were collected onto slides and successively mounted with Eukitt (O. Kindler, Freiburg, Germany) or Aqua-Polymount (cat. No. 18606, Polysciences Inc., Warrington, PA, USA). St. 42 tadpoles were fixed for 1 h in 4% paraformaldehyde at room temperature, cryoprotected in 25% sucrose in phosphate buffer saline overnight, sectioned using a cryostat (12 µm-thick sections) and finally collected onto polarised slides. In accordance with published methods (D'Autilia et al., 2010), sections were subjected to ISH and then mounted with Aqua-Polymount. Images from hybridized sections were acquired by a Nikon Eclipse Ti microscope, connected to a Nikon DS-Ri3 digital camera and equipped with the software NIS-Elements AR 5.11.03. To prepare all image panels shown in this work, the software Adobe Photoshop CS 8.0.1 was used.
Supplementary Material
Acknowledgements
The authors thank Salvatore Di Maria for frog care. This research was funded by intramural grants from University of Pisa to MA.
References
Argente-Escrig H., Vílchez J. J., Frasquet M., Muelas N., Azorín I., Vílchez R., Millet-Sancho E., Pitarch I., Tomás-Vila M., Vázquez-Costa J. F., Mas-Estellés F., Marco-Marín C., Espinós C., Serrano-Lorenzo P., Martin M. A., Lupo V., Sevilla T. (2022). A novel TRMT5 mutation causes a complex inherited neuropathy syndrome: The role of nerve pathology in defining a demyelinating neuropathy . Neuropathology and Applied Neurobiology 48: e12817.
Balke D., Kuss A., Müller S. (2015). Landmarks in the Evolution of (t)-RNAs from the Origin of Life up to Their Present Role in Human Cognition. Life 6: 1.
Bhreathnach U., Griffin B., Brennan E., Ewart L., Higgins D., Murphy M. (2017). Profibrotic IHG-1 complexes with renal disease associated HSPA5 and TRAP1 in mitochondria. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1863: 896-906.
Blaze J., Akbarian S. (2022). The tRNA regulome in neurodevelopmental and neuropsychiatric disease. Molecular Psychiatry 27: 3204-3213.
Bruna de Lima C., Cristina dos Santos É, Sirard M. A. (2023). DOHaD: A Menagerie of Adaptations and Perspectives: The interplay between early embryo metabolism and mitoepigenetic programming of development. Reproduction 166: F15-F26.
Chakraborty P. K., Schmitz-Abe K., Kennedy E. K., Mamady H., Naas T., Durie D., Campagna D. R., Lau A., Sendamarai A. K., Wiseman D. H., May A., Jolles S., Connor P., Powell C., Heeney M. M., Giardina P.J., Klaassen R. J., Kannengiesser C., Thuret I., Thompson A. A., Marques L., Hughes S., Bonney D. K., Bottomley S. S., Wynn R. F., Laxer R. M., Minniti C. P., Moppett J., Bordon V., Geraghty M., Joyce P. B. M., Markianos K., Rudner A. D., Holcik M., Fleming M. D. (2014). Mutations in TRNT1 cause congenital sideroblastic anemia with immunodeficiency, fevers, and developmental delay (SIFD). Blood 124: 2867-2871.
Chen D., Li F., Yang Q., Tian M., Zhang Z., Zhang Q., Chen Y., Guan M.X. (2016). The defective expression of gtpbp3 related to tRNA modification alters the mitochondrial function and development of zebrafish. The International Journal of Biochemistry & Cell Biology 77: 1-9.
Chen X., Ji Y., Liu R., Zhu X., Wang K., Yang X., Liu B., Gao Z., Huang Y., Shen Y., Liu H., Sun H. (2023). Mitochondrial dysfunction: roles in skeletal muscle atrophy. Journal of Translational Medicine 21: 503.
Chen Y., Yuen W. H., Fu J., Huang G., Melendez A. J., Ibrahim F. B. M., Lu H., Cao X. (2007). The Mitochondrial Respiratory Chain Controls Intracellular Calcium Signaling and NFAT Activity Essential for Heart Formation in Xenopus laevis . Molecular and Cellular Biology 27: 6420-6432.
Corcoran J. B., McCarthy S., Griffin B., Gaffney A., Bhreathnach U., Börgeson E., Hickey F. B., Docherty N. G., Higgins D. F., Furlong F., Martin F., Godson C., Murphy M. (2013). IHG-1 must be localised to mitochondria to decrease Smad7 expression and amplify TGF-β1-induced fibrotic responses. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1833: 1969-1978.
D'Autilia S., Broccoli V., Barsacchi G., Andreazzoli M. (2010). Xenopus Bsx links daily cell cycle rhythms and pineal photoreceptor fate . Proceedings of the National Academy of Sciences 107: 6352-6357.
DeLuca A. P., Whitmore S. S., Barnes J., Sharma T. P., Westfall T. A., Scott C. A., Weed M. C., Wiley J. S., Wiley L. A., Johnston R. M., Schnieders M. J., Lentz S. R., Tucker B. A., Mullins R. F., Scheetz T. E., Stone E. M., Slusarski D. C. (2016). Hypomorphic mutations in TRNT1 cause retinitis pigmentosa with erythrocytic microcytosis . Human Molecular Genetics 25: 44-56.
Edvardson S., Elbaz-Alon Y., Jalas C., Matlock A., Patel K., Labbé K., Shaag A., Jackman J. E., Elpeleg O. (2016). A mutation in the THG1L gene in a family with cerebellar ataxia and developmental delay. neurogenetics 17: 219-225.
Fisher M., James-Zorn C., Ponferrada V., Bell A. J., Sundararaj N., Segerdell E., Chaturvedi P., Bayyari N., Chu S., Pells T., Lotay V., Agalakov S., Wang D. Z., Arshinoff B. I., Foley S., Karimi K., Vize P. D., Zorn A. M., (2023). Xenbase: key features and resources of the Xenopus model organism knowledgebase. Genetics 224: iyad018.
Giannaccini M., Giudetti G., Biasci D., Mariotti S., Martini D., Barsacchi G., Andreazzoli M. (2013). Brief Report: Rx1 Defines Retinal Precursor Identity by Repressing Alternative Fates Through the Activation of TLE2 and Hes4. Stem Cells 31: 2842-2847.
Giudetti G., Giannaccini M., Biasci D., Mariotti S., Degl'innocenti A., Perrotta M., Barsacchi G., Andreazzoli M. (2014). Characterization of the Rx1-dependent transcriptome during early retinal development. Developmental Dynamics 243: 1352-1361.
Guo D., Hu K., Lei Y., Wang Y., Ma T., He D. (2004). Identification and Characterization of a Novel Cytoplasm Protein ICF45 That Is Involved in Cell Cycle Regulation. Journal of Biological Chemistry 279: 53498-53505.
Han R., Chu M., Gao J., Wang J., Wang M., Ma Y., Jia T., Zhang X. (2023). Compound heterozygous variants of THG1L result in autosomal recessive cerebellar ataxia. Journal of Human Genetics 68: 843-848.
Han Y., Ishibashi S., Iglesias-Gonzalez J., Chen Y., Love N. R., Amaya E. (2018). Ca2+-Induced Mitochondrial ROS Regulate the Early Embryonic Cell Cycle. Cell Reports 22: 218-231.
Hickey F. B., Corcoran J. B., Docherty N. G., Griffin B., Bhreathnach U., Furlong F., Martin F., Godson C., Murphy M. (2011). IHG-1 Promotes Mitochondrial Biogenesis by Stabilizing PGC-1α. Journal of the American Society of Nephrology 22: 1475-1485.
Hickey F. B., Corcoran J. B., Griffin B., Bhreathnach U., Mortiboys H., Reid H. M., Andrews D., Byrne S., Furlong F., Martin F., Godson C., Murphy M. (2014). IHG-1 Increases Mitochondrial Fusion and Bioenergetic Function. Diabetes 63: 4314-4325.
Klymkowsky M.W. (2011). Mitochondrial activity, embryogenesis, and the dialogue between the big and little brains of the cell. Mitochondrion 11: 814-819.
Martini D., Digregorio M., Voto I. A. P., Morabito G., Degl'Innocenti A., Giudetti G., Giannaccini M., Andreazzoli M. (2023). Kdm7a expression is spatiotemporally regulated in developing Xenopus laevis embryos, and its overexpression influences late retinal development . Developmental Dynamics 253: 508-518.
May-Panloup P., Boguenet M., El Hachem H., Bouet P.E., Reynier P. (2021). Embryo and Its Mitochondria. Antioxidants 10: 139.
Messina A., Lan L., Incitti T., Bozza A., Andreazzoli M., Vignali R., Cremisi F., Bozzi Y., Casarosa S. (2015). Noggin-Mediated Retinal Induction Reveals a Novel Interplay Between Bone Morphogenetic Protein Inhibition, Transforming Growth Factor β, and Sonic Hedgehog Signaling. Stem Cells 33: 2496-2508.
Mughal B. B., Leemans M., Spirhanzlova P., Demeneix B., Fini J.B. (2018). Reference gene identification and validation for quantitative real-time PCR studies in developing Xenopus laevis. Scientific Reports 8: 496.
Murphy M., Docherty N. G., Griffin B., Howlin J., McArdle E., McMahon R., Schmid H., Kretzler M., Droguett A., Mezzano S., Brady H. R., Furlong F., Godson C., Martin F. (2008). IHG-1 Amplifies TGF-β1 Signaling and Is Increased in Renal Fibrosis. Journal of the American Society of Nephrology 19: 1672-1680.
Murphy M., Hickey F., Godson C. (2013). IHG-1 amplifies TGF-β1 signalling and mitochondrial biogenesis and is increased in diabetic kidney disease. Current Opinion in Nephrology and Hypertension 22: 77-84.
Newport J., Kirschner M. (1982). A major developmental transition in early xenopus embryos: II. control of the onset of transcription. Cell 30: 687-696.
Phizicky E. M., Hopper A. K. (2023). The life and times of a tRNA. RNA 29: 898-957.
Rabin R., Hirsch Y., Johansson M. M., Ekstein J., Ekstein A., Pappas J. (2021). Severe epileptic encephalopathy associated with compound heterozygosity of THG1L variants in the Ashkenazi Jewish population . American Journal of Medical Genetics Part A 185: 1589-1597.
Reinhart M., Muraresku C., Ganetzky R., (1993). TRMU Deficiency. In GeneReviews®. (Ed. Adam M. P., Feldman J., Mirzaa G. M., Pagon R. A., Wallace S. E., Bean L. J. H., Gripp K. W., Amemiya A., ) University of Washington, Seattle, Seattle.
Rosen A. E., Brooks B. S., Guth E., Francklyn C. S., Musier-Forsyth K. (2006). Evolutionary conservation of a functionally important backbone phosphate group critical for aminoacylation of histidine tRNAs. RNA 12: 1315-1322.
Sasarman F., Thiffault I., Weraarpachai W., Salomon S., Maftei C., Gauthier J., Ellazam B., Webb N., Antonicka H., Janer A., Brunel-Guitton C., Elpeleg O., Mitchell G., Shoubridge E. A. (2015). The 3′ addition of CCA to mitochondrial tRNASer(AGY) is specifically impaired in patients with mutations in the tRNA nucleotidyl transferase TRNT1. Human Molecular Genetics 24: 2841-2847.
Schulz R., Schlüter K.D. (2023). Importance of Mitochondria in Cardiac Pathologies: Focus on Uncoupling Proteins and Monoamine Oxidases. International Journal of Molecular Sciences 24: 6459.
Session A. M., Uno Y., Kwon T., Chapman J. A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., van Heeringen S. J., Quigley I., Heinz S., Ogino H., Ochi H., Hellsten U., Lyons J. B., Simakov O., Putnam N., Stites J., Kuroki Y., Tanaka T., Michiue T., Watanabe M., Bogdanovic O., Lister R., Georgiou G., Paranjpe S. S., van Kruijsbergen I., Shu S., Carlson J., Kinoshita T., Ohta Y., Mawaribuchi S., Jenkins J., Grimwood J., Schmutz J., Mitros T., Mozaffari S. V., Suzuki Y., Haramoto Y., Yamamoto T. S., Takagi C., Heald R., Miller K., Haudenschild C., Kitzman J., Nakayama T., Izutsu Y., Robert J., Fortriede J., Burns K., Lotay V., Karimi K., Yasuoka Y., Dichmann D. S., Flajnik M. F., Houston D. W., Shendure J., DuPasquier L., Vize P. D., Zorn A. M., Ito M., Marcotte E. M., Wallingford J. B., Ito Y., Asashima M., Ueno N., Matsuda Y., Veenstra G. J. C., Fujiyama A., Harland R. M., Taira M., Rokhsar D. S. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336-343.
Shaheen R., Maddirevula S., Ewida N., Alsahli S., Abdel-Salam G. M.H., Zaki M. S., Tala S. A., Alhashem A., Softah A., Al-Owain M., Alazami A. M., Abadel B., Patel N., Al-Sheddi T., Alomar R., Alobeid E., Ibrahim N., Hashem M., Abdulwahab F., Hamad M., Tabarki B., Alwadei A. H., Alhazzani F., Bashiri F. A., Kentab A., Şahintürk S., Sherr E., Fregeau B., Sogati S., Alshahwan S. A. M., Alkhalifi S., Alhumaidi Z., Temtamy S., Aglan M., Otaify G., Girisha K. M., Tulbah M., Seidahmed M. Z., Salih M. A., Abouelhoda M., Momin A. A., Saffar M. A., Partlow J. N., Arold S. T., Faqeih E., Walsh C., Alkuraya F. S. (2019). Genomic and phenotypic delineation of congenital microcephaly. Genetics in Medicine 21: 545-552.
Skamagki M., Wicher K. B., Jedrusik A., Ganguly S., Zernicka-Goetz M. (2013). Asymmetric Localization of Cdx2 mRNA during the First Cell-Fate Decision in Early Mouse Development. Cell Reports 3: 442-457.
Terada K., Kitayama A., Kanamoto T., Ueno N., Furukawa T. (2006). Nucleosome regulator Xhmgb3 is required for cell proliferation of the eye and brain as a downstream target of Xenopus rax/Rx1. Developmental Biology 291: 398-412.
(2004). Fast Release Clones: A High Throughput Expression Analysis. ZFIN Direct Data Submission.. https://zfin.org/ZDB-PUB-040907-1
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3: RESEARCH0034.
Walker M. A., Lerman-Sagie T., Swoboda K., Lev D., Blumkin L. (2019). Refining the phenotype of the THG1L (p.Val55Ala mutation)-related mitochondrial autosomal recessive congenital cerebellar ataxia . American Journal of Medical Genetics Part A 179: 1575-1579.
White R. J., Collins J. E., Sealy I. M., Wali N., Dooley C. M., Digby Z., Stemple D. L., Murphy D. N., Billis K., Hourlier T., Füllgrabe A., Davis M. P., Enright A. J., Busch-Nentwich E. M., (2017). A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife 6: .
Wiseman D. H., May A., Jolles S., Connor P., Powell C., Heeney M. M., Giardina P. J., Klaassen R. J., Chakraborty P., Geraghty M. T., Major-Cook N., Kannengiesser C., Thuret I., Thompson A. A., Marques L., Hughes S., Bonney D. K., Bottomley S. S., Fleming M. D., Wynn R. F. (2013). A novel syndrome of congenital sideroblastic anemia, B-cell immunodeficiency, periodic fevers, and developmental delay (SIFD). Blood 122: 112-123.
Yanai I., Peshkin L., Jorgensen P., Kirschner M. W. (2011). Mapping Gene Expression in Two Xenopus Species: Evolutionary Constraints and Developmental Flexibility. Developmental Cell 20: 483-496.
Zhang Q., Ouyang Q., Xiang J., Li H., Lv H., An Y. (2023). Pathogenicity Analysis of a Novel Variant in GTPBP3 Causing Mitochondrial Disease and Systematic Literature Review. Genes 14: 552.
Zhang Q., Zhang L., Chen D., He X., Yao S., Zhang Z., Chen Y., Guan M. X., (2018). Deletion of Mtu1 (Trmu) in zebrafish revealed the essential role of tRNA modification in mitochondrial biogenesis and hearing function. Nucleic acids research 46: 10930-10945.