Int. J. Dev. Biol. 66: 373 - 381 (2022)
Cyclooxygenase-2 plays a crucial role during myocardial patterning of developing chick
Original Article | Published: 22 December 2022
Abstract
Cyclooxygenase-2 (COX-2), a member of the Cyclooxygenase family, initiates the biosynthesis of prostanoids that regulates various cellular functions. Our pilot attempt revealed that the administration of etoricoxib, an inhibitor specific for COX-2, induces abnormal looping in the chicken heart. The present study attempts to reveal the mechanistic details of etoricoxib-induced abnormal cardiac looping. The activity of COX-2 was inhibited by administering 3.5 μg of etoricoxib into the egg’s air cell on day zero of incubation. The gene and protein expression patterns of key mediators of heart development were then analyzed on day 2 (HH12) and day 3 (HH20). Reduced COX-2 activity altered the expressions of upstream regulators of organogenesis like Wnt11, BMP4, and SHH in the etoricoxib-exposed embryos. The observed expression shifts in the downstream regulators of myocardial patterning (MYOCD, HAND2, GATA4, GATA5, and GATA6) in the treated embryos corroborate the above results. In addition, the reduction in COX-2 activity hampered cardiomyocyte proliferation with a concomitant increase in the apoptosis rate. In conclusion, the collective effect of altered expression of signaling molecules of myocardial patterning and compromised cardiomyocyte turnover rate could be the reason behind the looping defects observed in the heart of etoricoxib-treated chick embryos.
Keywords
chick embryo, heart development, cyclooxygenase-2, looping, trabeculation
Introduction
Congenital malformations are observed in at least 1% of newborn babies and conotruncal heart defects pose the highest in-utero mortality risks (Lutin et al., 1999; Dees et al., 2000). There is ample evidence suggesting that mortality remains high even after postnatal surgical repair (Khoshnood et al., 2012), making it imperative to understand the genesis of these defects.
Research on heart development has witnessed a renaissance in recent years. We still do not understand the defects’ correlation with the looping, convergence, and wedging process. As the segments of the cardiac tube are in sequence and are not separated, looping creates an inflow and an outflow tract (proximal and distal tract) (Van den Hoff et al., 1999). Both inflow and outflow tracts are brought together craniocaudally through convergence (Yelbuz et al., 2002). After the tract has converged, the inflow portion of the tube nestles behind the ventrally placed outflow tract, beginning a new period of outflow tract adjustment called wedging (Bartelings and Gittenberger-de, 1989). These processes occur in heart tubes and develop into a four-chambered heart in aves and mammals. In aves, the heart tube development starts at day one, and due to external development, looping patterns are evident in chick embryos of day 2 and day 3. Further, the molecular signaling was also reported to be similar in the case of aves and mammals, making the chick embryo an excellent model to study heart development (Olson, 2006).
Convergence and wedging of the heart tube require multiple cellular events such as cell proliferation, migration, and differentiation governed by molecules such as Wnts, BMPs, GATAs, and Shh. The expression of zinc finger transcription factors (GATA4, GATA5, and GATA6), which are known to regulate cell differentiation and cell number in the heart field, has been reported to be conserved throughout the vertebrate lineage (Nemer, 2008; Van Berlo and Maillet, 2010). During the initiation of the heart tube, myocardin expresses in the cells destined to become cardiac muscle (Chen et al., 2008). Moreover, HAND2 guides the migrating cells of the heart to achieve symmetry during the looping process (Laurent et al., 2017). Studies suggested that T-box transcription factors (TBX) interact with GATA genes, which coherently help sculpt vertebrates’ hearts (Stennard et al., 2003). GATA5 assists in regular cardiomyocyte migration, especially during ventricular wall compaction. Trabeculation defects and valvular dysfunction have been identified as consequences of deviated GATA5 expression in chick embryos (Pérez-Pomares and Pompa, 2011).
Among the variety of regulatory factors regulating cardiac tube looping and cardiac patterning, a key molecule is COX-2, an inducible isoform of cyclooxygenase that catalyzes the formation of PGE2 from arachidonic acid (Ricciotti and FitzGerald, 2011). COX-2 mediated PGE2 synthesis plays a crucial role in cellular events such as proliferation, migration, and differentiation by modulating the regulators of signaling pathways such as Wnt/β-catenin, BMP, and TGF-β (Neil et al., 2008; Jana et al., 2016; Buch et al., 2017). A study by Xu and coworkers showed that COX-2 inhibition using celecoxib causes defective heart formation and hinders the looping of heart tubes in zebrafish embryos (Xu et al., 2011).
A previous study from our laboratory showed that COX-2 inhibition by etoricoxib causes defects in limb development, vascularization, tissue integrity, and organ patterning in developing chicks, where the most lethal malformation was found in the heart region (Verma et al., 2021). This study observed that selective COX-2 inhibition reduces levels of PGE2 while the remaining prostanoids maintain their average titer. Based on these findings and other reported interactions of COX-2 with different signaling molecules, we hypothesize that the pathways regulating heart tube looping and chamber formation might be affected due to COX-2 inhibition, resulting in developmental heart defects in embryos.
In order to validate this notion, the present study inhibited COX-2 activity using etoricoxib, a selective COX-2 inhibitor, and its effects on the regulators of heart tube looping and chamber formation were ascertained in day 2 and day 3 chick embryos.
Results
Defective looping of heart tube on HH12 (day 2)
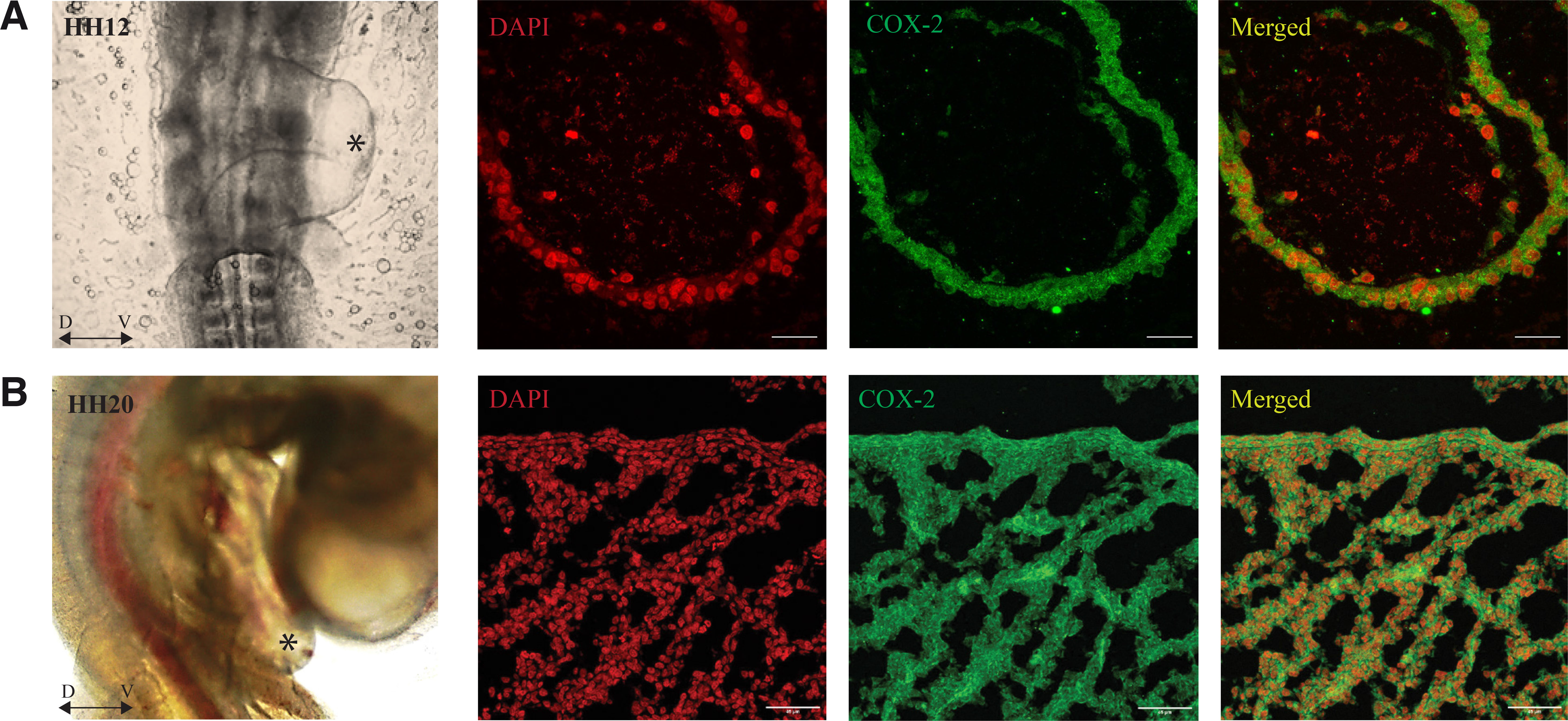
Immunolocalization indicates that COX-2 is located in the cytoplasmic region of the heart tube cells (Fig. 1A).
Fig. 1. Immunolocalization of COX-2 in the developing heart of control embryos.
(A) HH12 (day 2) control embryo (ventral side) shows COX-2 localization in the cytoplasm of a single layer of cardiomyocytes of the developing heart tube. The black asterisk indicates the section plane of the heart region. (B) HH20 (day 3) control embryo (ventral side) shows COX-2 protein expression in the cytoplasm of cells on the outer wall of the heart; also, the trabecula shows the presence of COX-2. The black asterisk shows the plane of section of the heart region, D ↔ V: represent Dorsal-Ventral orientation.
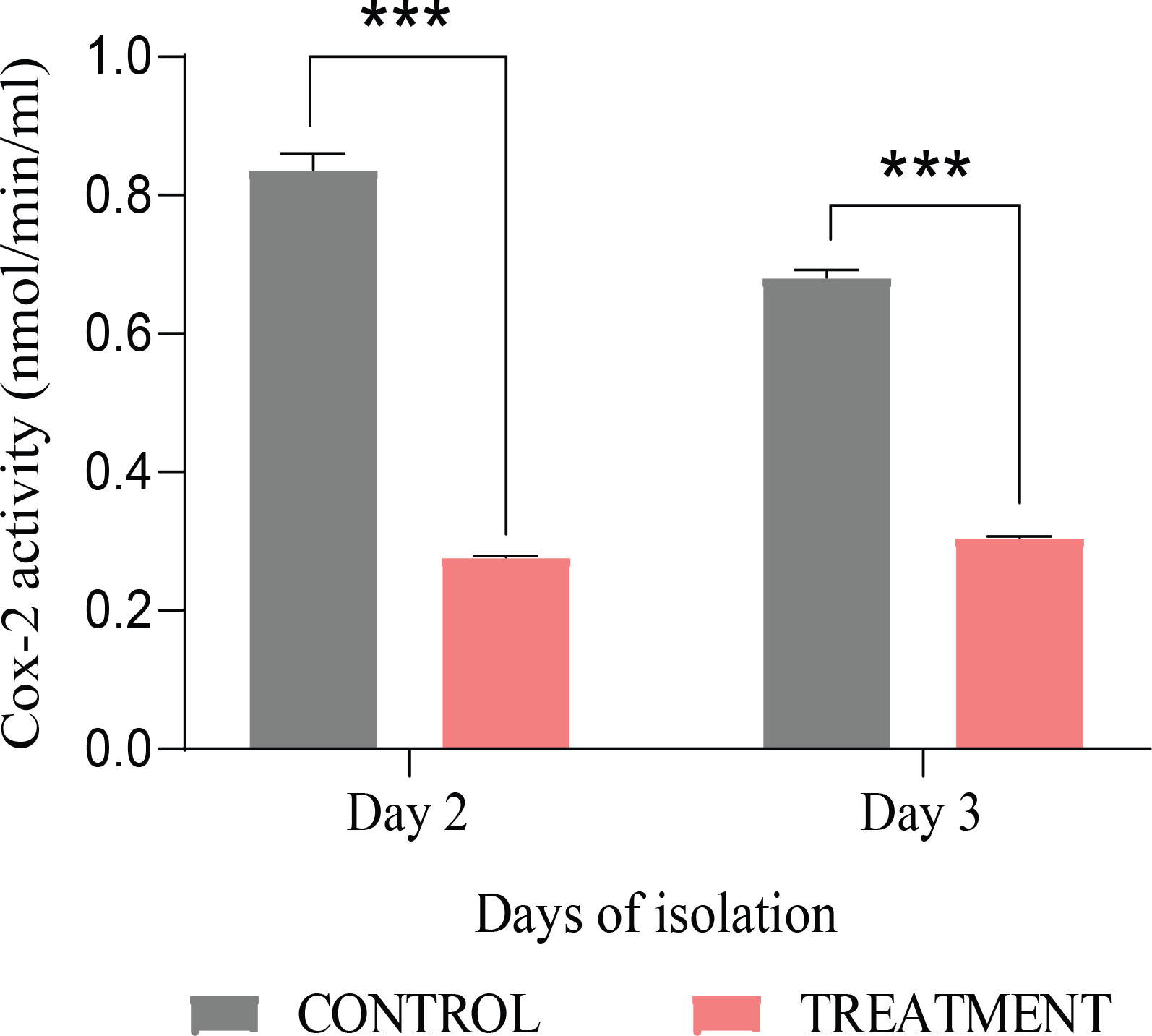
COX-2 activity was recorded in control and etoricoxib-treated embryos. There was a significant decrease in the COX-2 activity on day 2 of the developing embryo (Fig. 2, Table S1).
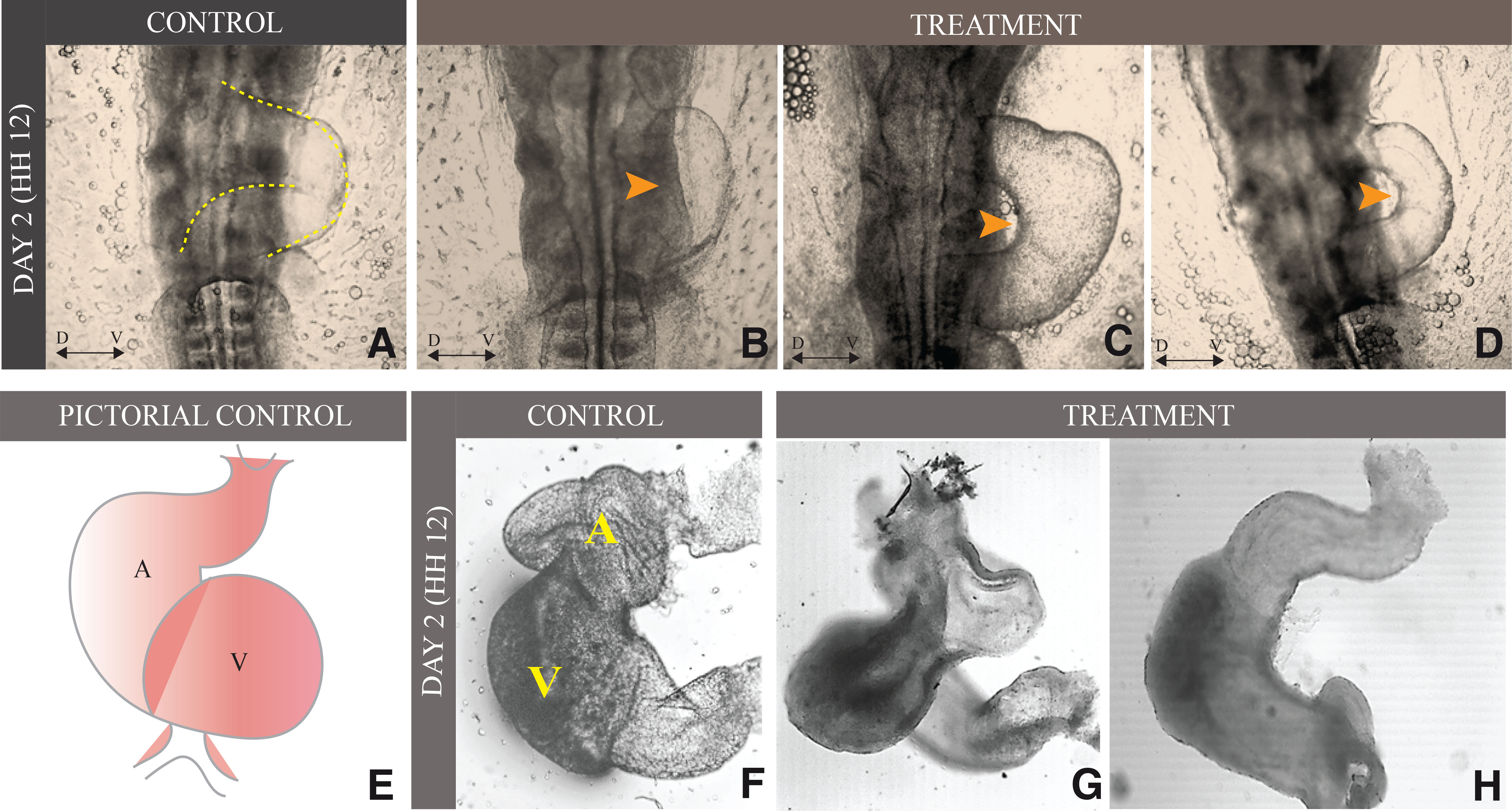
On day 2 (HH 12), the control group embryo showed a typical S-shaped heart tube (Fig. 3A). In contrast, the embryos from the treatment group showed obvious signs of defective heart looping. In addition, the rate of heart tube loop malformations was significantly high in the treated embryos. Aberrations, such as underdeveloped heart, distorted heart tube, and defective heart tube curvature, were frequently observed in etoricoxib-treated embryos (Fig. 3B-D). A visual comparison of heart tissues isolated from the control and treatment group of day 2 embryos suggests heart-loop defect (Fig. 3F-H).
Fig. 3. Etoricoxib exposure causes abnormal heart tube looping in the embryo on day 2.
(A) Normal looping of the heart tube found in the control group of the embryo on the ventral side (the dotted line represents normal looping of the heart).(B-D) Abnormal looping of heart tube in etoricoxib-treated embryos represented by orange arrowheads. (E) Pictorial representation of day two heart of control embryo.(F) Isolated heart of control embryo.(G-H) Abnormal looping in isolated heart tube and uneven thickening of the tube. A: Auricle, V: Ventricle, D ↔ V: represent Dorsal-Ventral orientation.
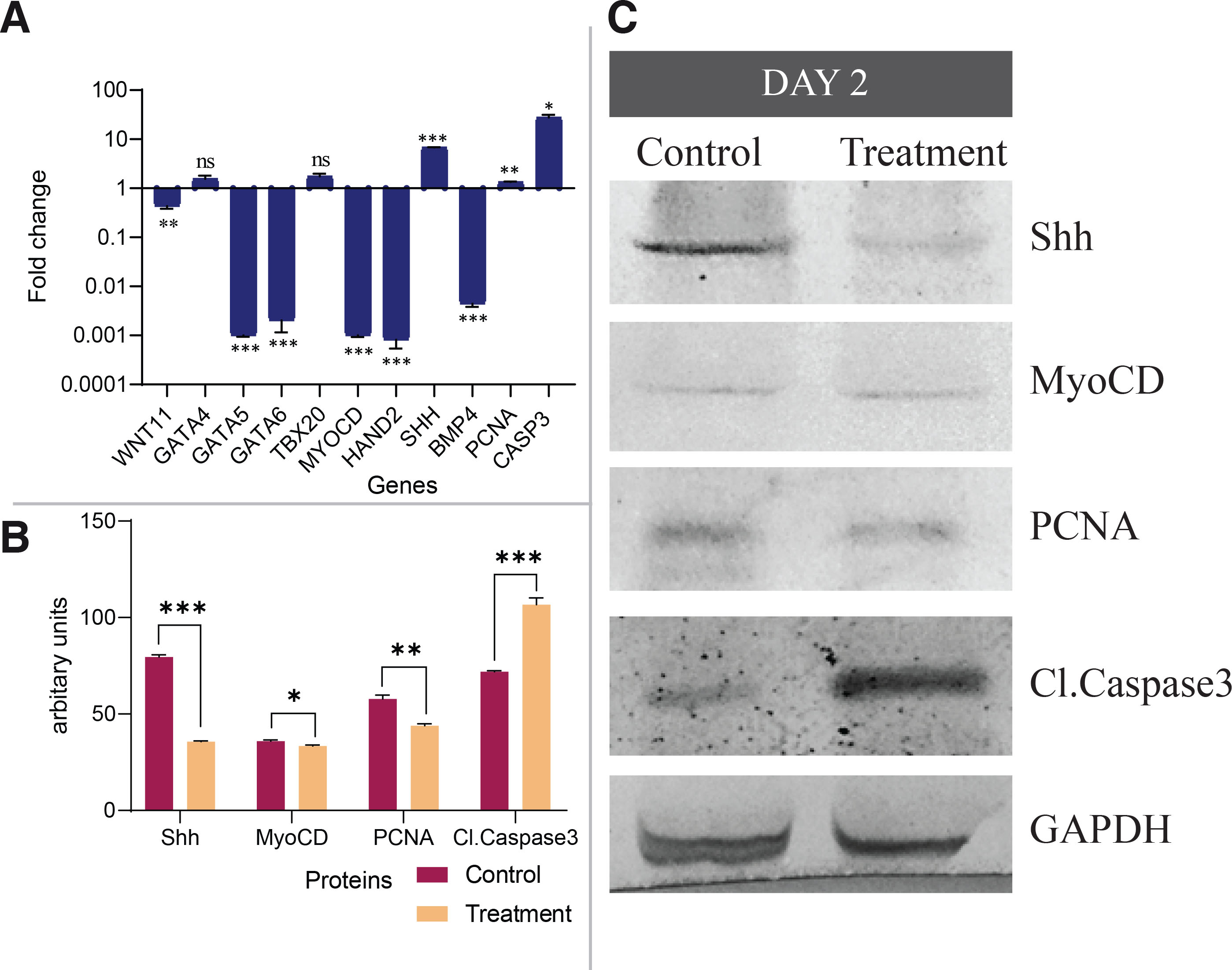
Studies involving gene expression were carried out to determine heart-loop deviation. The mRNA levels of genes that play a significant role in heart tube patterning and looping, viz. WNT11, GATA5, GATA6, and BMP4 were significantly reduced in the etoricoxib-treated embryos (Fig. 4A). However, the transcript levels of genes involved in the looping and maturation of the cardiomyocytes showed an aberrant expression pattern under the ablation of COX-2 activity. SHH, PCNA, GATA4, TBX20, and CASP3 mRNA levels increased, while those of HAND2 decreased in the treated embryo group compared to control embryos. Relative mRNA expression levels of the cardiomyocyte-specific gene, namely MYOCD, also showed a significant reduction under the etoricoxib treatment (Fig. 4A, Table S2).
Fig. 4. Expression pattern of genes and protein regulating the looping of heart tube of day two embryos.
(A) mRNA levels of various genes of cardiogenesis. Fold changes are expressed as Mean±SEM. Fold change values for the control embryo is 1.0 for all the genes, n=3 with 30 eggs per group per day; *p≤0.05; **p≤0.01; ***p≤0.001. (B) Quantification of western blots by densitometry. Relative band intensities were normalized to the intensity of GAPDH of the respective sample; *p≤0.05; **p≤0.01; ***p≤0.001. (C) Protein levels of Shh, MyoCD, PCNA, and Cl.Caspase3. GAPDH was taken as a loading control.
Concomitant immunoblot analysis for protein revealed reduced Shh, MyoCD, PCNA, and Cl.Caspase3 levels in treated embryos on day 2 compared to control embryos (Fig. 4 B,C, Table S4).
Aberration in heart chamber formation on HH 20 (day 3)
Immunolocalization of COX-2 showed its expression in a normally developed day three embryo. The expression of COX-2 was found in the inner wall of the developing heart tube and in the trabeculae region (Fig. 1B).
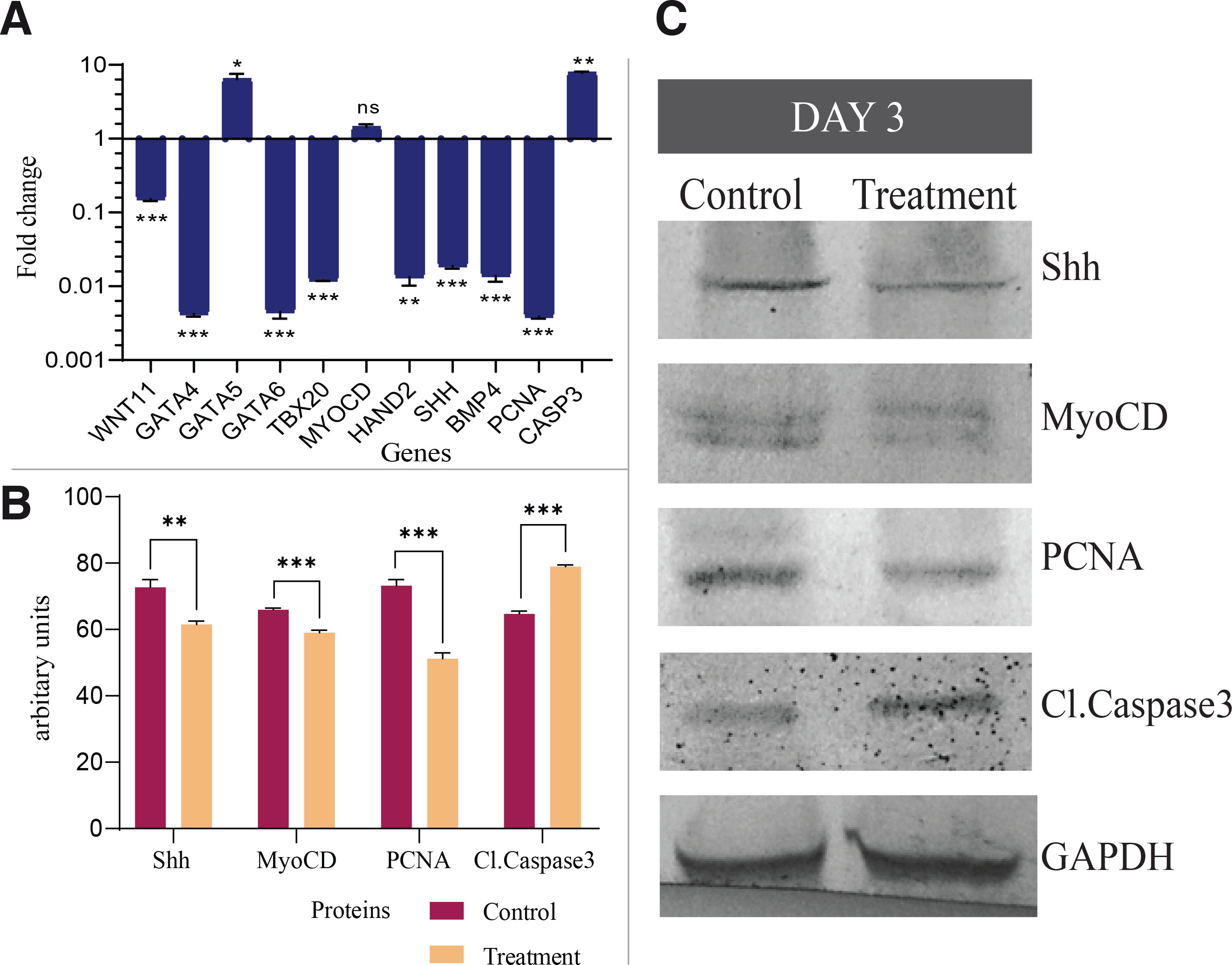
Embryos from the day three treated group (HH 20) showed a significant decrease in COX-2 activity (Fig. 2) accompanied by a high rate of heart abnormalities. The control group displayed normal development and achieved the characteristic conical shape with heart tubes differentiated into auricles and ventricles (Fig. 5A). In contrast, the treated embryos showed many defects, including abnormal bending of the heart tube and straight heart tube. Moreover, the heart chambers, atria, and ventricles are not fully formed in treated embryos compared to control embryos (Fig. 5 B,D). Key signaling molecules involved in heart patterning and functioning were investigated at transcript and protein levels to corroborate the findings. In day three treated embryos, relative mRNA expression levels exhibited remarkable downregulation of WNT11, GATA4, GATA6, TBX20, and BMP4. Likewise, the PCNA, SHH, and HAND2 transcript levels were conspicuously reduced in treated embryos (Fig. 6A, Table S3).
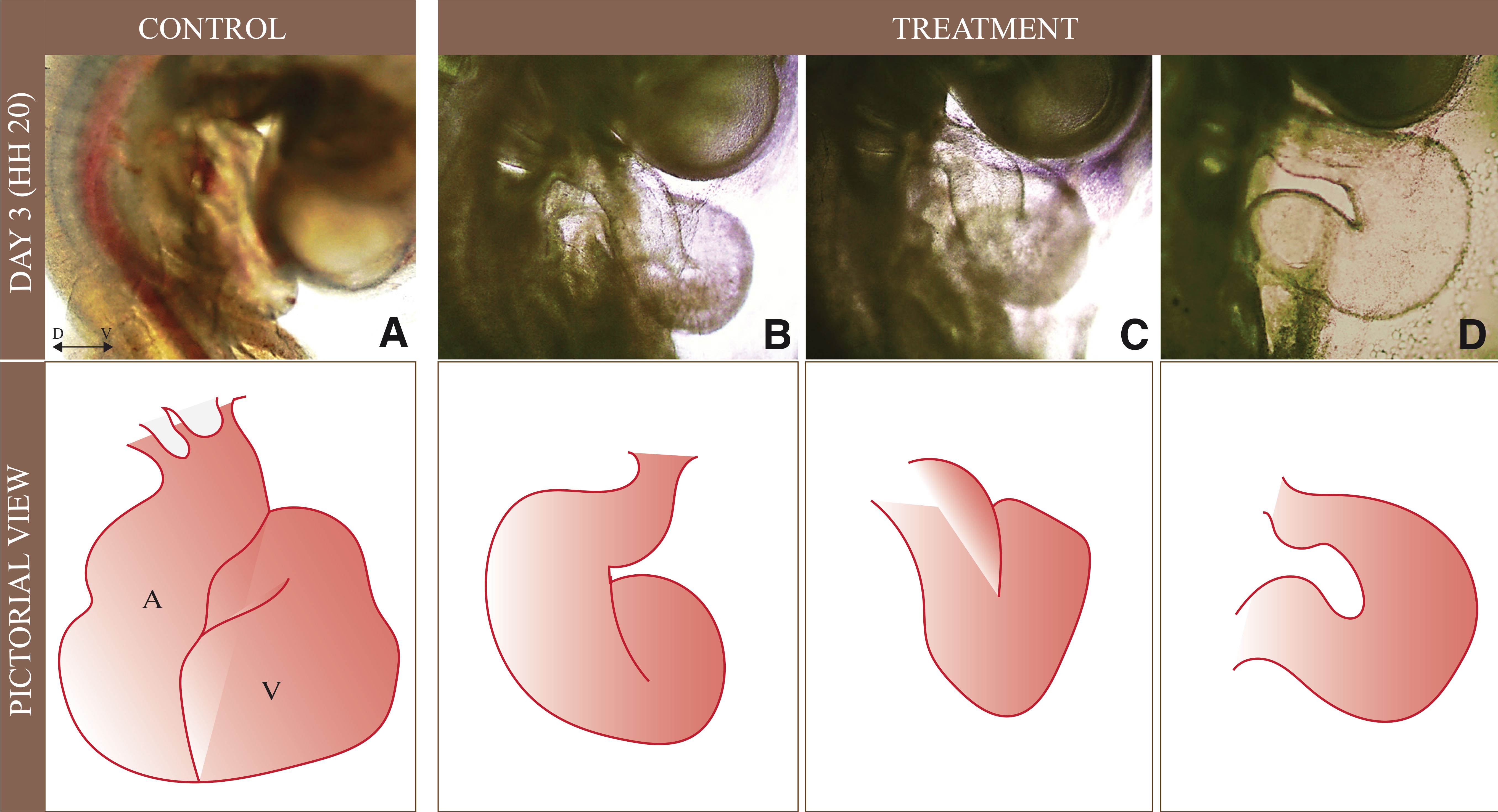
Fig. 5. Abnormal heart tube thickening and differentiation of chambers in the day three embryo.
(A) Control embryo has thickening at ventricle walls, (B-D) Treatment group of the embryo has delayed development of the heart tube and uneven thickening of ventricular walls; the lower panel shows a pictorial representation of day three heart for normally developing embryo and under the exposure of etoricoxib. A: Auricle, V: Ventricle, D ↔ V: represent Dorsal-Ventral orientation.
On the contrary, q-RT PCR analysis of GATA5, MYOCD, and CASP3 transcripts revealed an appreciable increase in their expression levels. To further substantiate the data at the transcript level, a Western blot was performed for key proteins responsible for cardiac patterning and loop formation. Densitometry analysis of Shh, MyoCD, and PCNA blots showed their levels significantly reduced under etoricoxib treatment. In contrast, Cl.Caspase3, an apoptotic marker, has exceptionally high expression levels in the embryos of the treatment group (Fig. 6 B,C, Table S4).
Fig. 6. Expression pattern of genes and protein regulating the heart tube thickening and chamber formation in day three embryos.
(A) mRNA levels of various genes of cardiogenesis. Fold changes are expressed as Mean±SEM. Fold change values for the control embryo is 1.0 for all the genes, n=3 with 30 eggs per group per day; ns-non significant; *p≤0.05; **p≤0.01; ***p≤0.001. (B) Spot densitometry of western blot. Relative band intensities were normalized to the intensity of GAPDH of the respective sample; ns-non significant; *p≤0.05; **p≤0.01; ***p≤0.001. (C) Protein levels of Shh, MyoCD, PCNA and Cl.Caspase3. GAPDH was taken as a loading control.
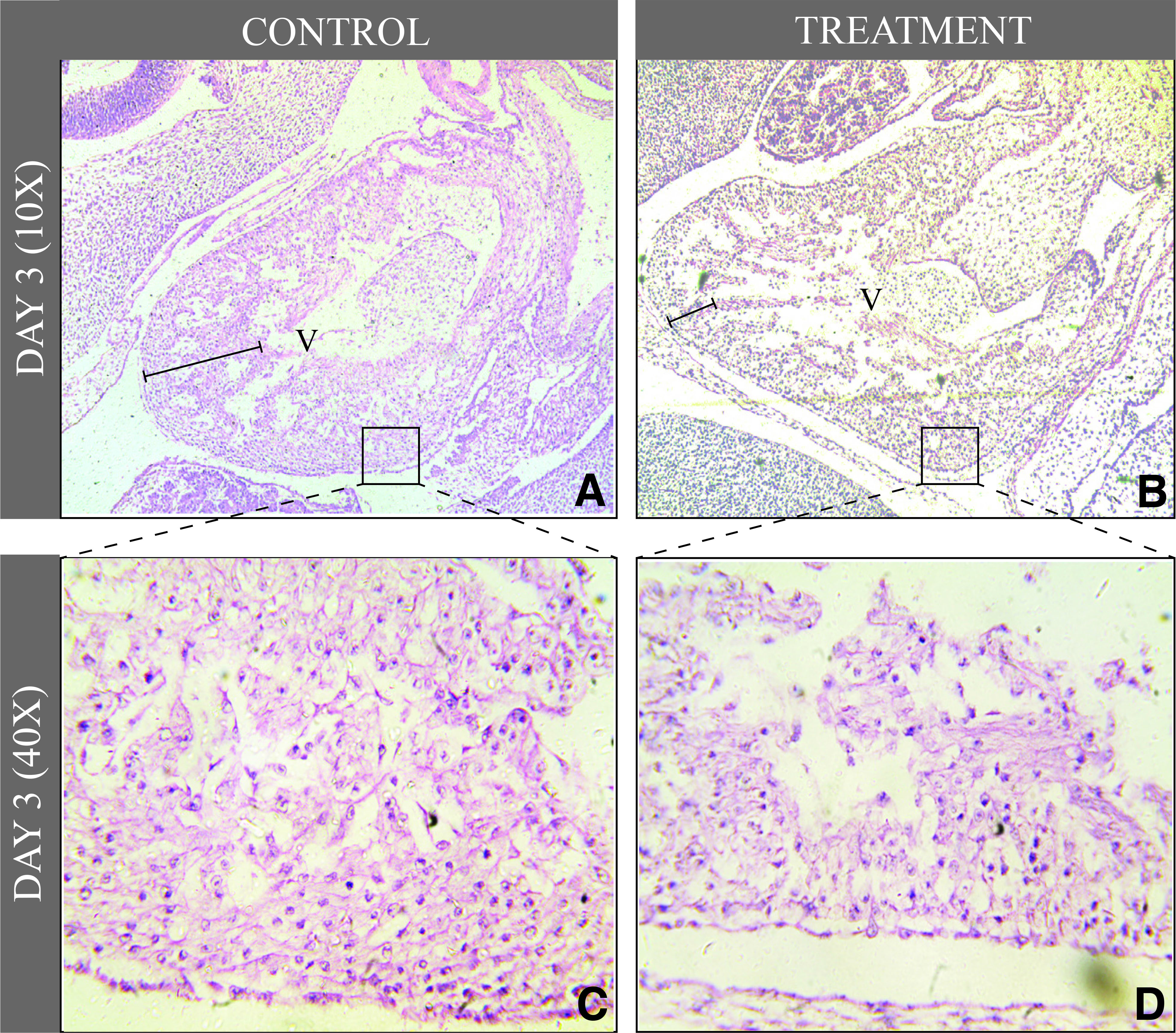
Additionally, the extent of heart patterning defects in HH 20 embryos was analyzed histologically. Control embryos revealed a functionally competent ventricular wall with standard compaction and trabeculation (Fig. 7 A,C). In opposition, the treatment group showed clear signs of ventricular non-compaction and defective trabeculation (Fig. 7 B,D).
Fig. 7. Histology of day three heart.
(A) Control heart (10X) has a triangular shape and dense ventricular walls at lower resolution (showed by the black line segment). (B) Etoricoxib treated heart have thin ventricular walls visible at 10x resolution (showed by the black line segment). (C) Inset image of control heart showing tightly packed cardiomyocytes showing normal trabeculation in the control group, magnification 40x. (D) Inset image of treatment group heart shows lack of cardiomyocytes packing in the ventricular wall. The loose arrangement of cells in the treated-embryonic heart confirmed the defect of trabeculation at the same stage as compared to the control group.
Discussion
Many studies in mammals and other animal models have shown that COX-2-derived PGE2 is an essential mediator of various physiological and pathological processes (Bondesen et al., 2006; Pakrasi and Jain, 2008; Nørregaard et al., 2015). Although studies have previously implicated the possible association of COX-2-derived PGE2 with an embryonic malformation (Randall et al., 1991), its role in the development of the heart or organogenesis of an embryo is still unknown. In this study, we hypothesized that aberrant COX-2 expression might interfere with embryological development during a critical period of embryogenesis, especially in heart development. Indeed, the COX-2 single knockout mice fetus exhibited severe defects in the axial skeleton and died shortly after birth due to respiratory distress (Shim et al., 2010). In a typically developing embryo, COX-2 expression is present to guide organogenesis, which was depicted by the immunolocalization method that clearly marked its contribution in the developing heart tube as well as walls of trabeculae on day 3.
The present study showed that when COX-2 activity has hampered, apoptosis in developing heart tubes dramatically increases, and the number of cardiomyocytes decreases. Additionally, trabeculae formation and cardiomyocytes condensation are impaired, leading to erroneous heart chamber formation under inhibition of COX-2 activity.
The Wnt signaling, both canonical and non-canonical, primarily regulates the heart tube’s initial pattering, which plays an important role in deciding the fate of cardiomyocytes (Pandur et al., 2002; Piven and Winata, 2017). In an isolated study, in a quail embryo, the dominant-negative Wnt11 construct decreased the expression of cardiac markers, hence not allowing cells to differentiate into cardiomyocytes in quail embryo. The expression of Wnt11 is found at a very early stage when paraxial plate mesoderm differentiates in developing chick embryos (Eisenberg and Eisenberg, 1999). We observed a significant decrease in Wnt11 level in the heart-forming tissues of day two and three chick embryos subjected to the selective inhibitor of COX-2 (etoricoxib). Due to the decrease in Wnt11 levels in the treatment group of embryos, the downstream molecules, such as GATA family members, showed deviated expression patterns.
Furthermore, specific genes and proteins were analyzed to understand the molecular aspects of the defects mentioned above. GATA5 is one such transcription factor that regulates myocyte number during heart development. GATA5 decrease in day two embryos hampers the cardiomyocyte formation, while on day three, it increases, which can be an outcome of a feedback loop. Its role in migration during early embryogenesis is also known (Peterkin et al., 2007). Deviant expression of GATA5 was reported to induce trabeculation defects, valvular dysfunction, epicardia, and mesenchymal malformation in chick embryos (Chen and Fishman, 2000; Laforest et al., 2011). Our findings also revealed a similar trend in expressing GATA5 and the anatomical features in the day two and day three embryos. We found deformed trabeculae in the treated group heart on day 3. Based on the results highlighted above, it is prudent to presume that COX-2 inhibition modulates the expression of GATA5.
Another transcription factor that facilitates the migration of cardiomyocytes is GATA4. It is reported that GATA4 expresses during the early stages of heart development, from the migration of pre-cardiac cells to the complete heart tube formation (Lopez-Sanchez et al., 2009). It has been observed that in homozygous GATA4 null mice, the erroneous migration of pro-myocardial primordia resulted in two independent heart tubes (Molkentin et al., 1997). The experimental group of embryos showed impaired expression of GATA4 compared to the control embryos. The data shows that GATA4 increased on day two and then decreased on day three embryos under the etoricoxib treatment. Reports suggest that GATA4 and GATA5 genes cooperatively in zebrafish perform functions such as liver and heart organogenesis and myocyte population regulation. (Singh et al., 2010). There is ample evidence suggesting that the other GATA gene compensates for each GATA gene's expression if one gene goes downregulated (Laforest and Nemer., 2011). The antagonistic expression pattern observed for GATA4 and GATA5 for day two and day three treated embryos agrees with the above notion.
Recent studies demonstrated that conditional deletion of GATA6 leads to malformation of the cardiac outflow tract and aortic arch arteries (Lepore et al., 2006). In our studies, the level of the GATA6 transcription factor decreased on day two and day three under the treatment of etoricoxib. The declined levels of GATA6 led to defective outflow track formation in the day three embryo, which was visible in histology. Moreover, it has been suggested that increased levels of GATA6 may compensate for the loss of GATA4 in chicks (Narita et al., 1997). Similar results were also found in our studies on day three embryos when treated with etoricoxib.
In addition, a vital molecule responsible for cardiac morphogenesis is the T-box transcription factor (TBX). The TBX family members interact with GATA factors and, together, contribute to sculpting the heart in vertebrates. According to a report, when TBX20 mutates, heart development is seized at the tube stage with no further development (Plageman and Yutzey, 2004). Moreover, it has been reported that TBX20 co-express with GATA5 and regulates myocyte population (Laforest and Nemer, 2011). The morphological observations of day two embryos of the treatment group revealed looping defects in the heart tube. The transcript level analysis of TBX20 and GATA5 revealed an antagonistic expression pattern (heightened expression of TBX20 with a concomitant reduction in GATA5). It is well known that TBX20 acts as a transcriptional regulator of the ANF (atrial natriuretic factor). However, the promoter region of the ANF is responsive to other factors, one of which is TBX5.
Nevertheless, when TBX5 acts as an activator, TBX20 acts as a repressor for ANF (Plageman and Yutzey, 2004). This intricate relationship between T-box transcriptional factors and downstream genes allows the formation of inflow and outflow tracks, atrioventricular cushions and valves at different time points during cardiac morphogenesis (Brown et al., 2005). The lowered levels of TBX20 could hinder the standard sculpting of the heart in developing embryos despite having high levels of GATA5 under the etoricoxib treatment on day 3.
Moreover, the MYOCD gene that directs the myocardin protein synthesis expresses in an early embryo's heart field (Huang et al., 2009). The presence of myocardin is reported from the HH9 (day 1) stage of a developing chick when the cardiomyocyte-fated progenitors start to fuse until the valves and nodes of a developed heart are fully formed (Brand, 2003). MYOCD protein has also been reported to have many structural and functional roles in heart development, such as cell fusion, cardiomyocyte contractibility, and more. (Raphel et al., 2012). In the current study, COX-2 inhibition caused the downregulation of MYOCD compared to control in day two embryos. It has been informed that MYOCD overexpression reduces cell death and promotes tissue repair in a murine model of hindbrain ischemia (Madonna et al., 2013). Therefore, it is believed that reduced MYOCD expression could be an added reason behind the apoptosis and ameliorated morphogenesis in the developing heart tube under the ablated COX-2 activity in day two embryos.
Additionally, transcriptional factor HAND2 is been shown in the early heart field at HH16, and its downregulation leads to abnormal correct ventricle formation (Srivastava et al., 1997). It has also been reported that HAND2 moves to the outflow tract in the later stages of cardiac looping (Tsuchihashi et al., 2011). Here, treatment was observed to decrease HAND2 expression in embryonic heart tissue, which, together with a compromised level of MYOCD, would account for the looping defects noticed in the heart of day two and day three chick embryos.
SHH is another factor that regulates the right and left looping of the heart in early-developing embryos. It turns the heart tube in the right direction, further interacts with HAND, and leads to the maturation of the heart chambers (Tsuda et al., 1998). In our study, SHH gene expression increased on day two under the etoricoxib treatment, while on day three, its levels decreased as compared to respective controls. Deviant expression of the SHH gene leads to disruption in the looping pattern of the cardiac tube, which ultimately causes congenital heart defects in developing embryos. SHH signaling pathways are critical in regulating the normal proliferation and migration rate (Dyer and Kirby, 2009).
In this respect, the member of the TGF-β family BMP4 is also known to control cardiac cell proliferation and differentiation by interacting with Wnt (Ye et al., 2019). Reports showed that BMP4 provides the left-right integrity of heart structure in developing zebrafish embryos (Chen et al., 1997). The expression level of BMP4 was found to decrease in the treated group of day two and day three embryos. Subsided level of BMP4 suggests its reduced interaction with Wnt and their downstream molecules, leading to defective patterning of the heart tube.
The cardiomyocyte population has to be maintained for sculpting the heart tube. During heart tube development, the cardiomyocytes are in constantly proliferate (Linask et al., 2005). Initially, when a heart tube is formed, the apoptosis is found to be minimal; however, when the heart tube starts differentiating into chambers, the apoptosis rate in cardiomyocytes increases (Zhao and Rivkees, 2000). The ratio of proliferation and apoptosis maintains the cell number and progress of heart development. The level of PCNA in the etoricoxib-treated embryo decreased drastically, while that of Cl.Caspase3 increased, leading to fewer cells, as seen in the histology of heart sections.
Overall, due to COX-2 inhibition, a significant signaling molecule like Wnt11 was affected along with its downstream targets, such as GATAs and TBXs, which are interconnected for the heart tube patterning. Alteration in these might have resulted in the malformation of the heart in the chick embryo.
COX-2 inhibition resulted in heart tube looping alterations and defective trabeculation in the ventricular region of day two and day three chick embryos. The regulatory molecules were affected when levels of COX-2 went down. Alterations at gene and protein levels were reflected in the morphology of the developing heart. Our findings with chick embryos suggested that COX-2 has a crucial role in patterning the heart in vertebrates.
Materials and Methods
Animal procurement and maintenance
This study used the fertilized eggs of the Rhode Island Red (RIR) breed of domestic chicken. They were obtained from the Intensive Poultry Development Unit, Vadodara, Gujarat, India. Prior to incubation, eggs were candled to visualize the air sac and wiped with a povidone-iodine solution. All experiments were carried out per the committee’s guidelines for controlling and supervising experiments on animals (CPCSEA). The institutional animal ethics committee approved the protocols (IAEC No. MSU-Z/IAEC/09-2020).
Experiment design
Freshly laid eggs were categorized into two different groups, control, and treatment, randomly after disinfecting with povidone-iodine solution. All the experiments were performed thrice with 30 eggs in each group. Control and treatment groups of eggs were punctured at the marked air sac region with a sterile needle under the laminar airflow (LAF). In the control group of eggs, 50 µl of Mili-Q water was dispensed. For the treatment group, the same volume of 70 mg/ml (Low observed effective concentration-LOEC) etoricoxib prepared in Mili-Q water was administered. Thus, each treated embryo received 3.5 µg of etoricoxib dissolved in water.
All the experimental eggs were kept in an automated incubator at 37.5±0.5°C and relative humidity of 70-75% (Forma environmental chamber, Thermo Scientific USA). The eggs are automatically rotated every hour and incubated until the desired day embryos are isolated.
Embryo collection
Embryos were harvested on predetermined days, where their growth stages were confirmed by following Hamberger Hamilton (HH) stages of chick development (Hamburger and Hamilton, 1951). Embryos belonging to HH stage 12(day 2) and HH stage 20(day 3) were used for further experimentation and analysis.
Immunohistochemistry
The control embryos of day 2 (HH12) and day 3 (HH20) were fixed with 4% PFA at room temperature for 1 hour, followed by sucrose gradient then OCT was used for embedding. 10µm thick sections were taken through the heart region of the embryos using a cryostat (Reichert-Jung, cryo-cut). The sectioned embryos were rehydrated and washed with PBS, and then they were permeabilized at room temperature for 30 minutes and blocked. The embryo sections were blocked with Genei blocking solution for one hour (Genei Laboratories Private Limited, India). The sections were incubated at 4 °C overnight with rabbit anti-COX-2 antibody (1:500 dilution) (Sigma-Aldrich, USA, SAB4200576) and rabbit anti-Cl.Caspase3 antibody (1:500 dilution) (Sigma-Aldrich, USA, C8487) followed by secondary antibody goat anti-rabbit Alexa 488 (Invitrogen, USA, Cat no. A-11008) (1:1000 dilution) for one hour at room temperature in dark condition. Sections were incubated with nuclear stain DAPI (Invitrogen, USA, Cat no. D1306) for 10 minutes, followed by a wash with PBST and mounted on using Fluoromount. Imaging was done using Zeiss LSM-710 confocal microscope and analyzed in ImageJ software.
COX activity assay
The heart tissues were collected from both groups on day two and day three in 0.1 M Tris-EDTA buffer, and 10% homogenate of tissue was used to estimate COX-2 activity following the manufacturer's protocol (Cayman Chemical, USA).
A selective COX-1 inhibitor SC-560 was used for inhibitory wells. All the wells have assay buffer and heme along with samples (standard, background, and inhibitory wells). The plate was incubated for 10 minutes at room temperature, and 20 µl of the calorimetric substrate was added last. Absorbance was measured at 590 nm, which is the absorbance maxima of oxidized N,N,N′,N′-Tetramethyl-p-phenylenediamine (TMPD) corresponding amount of peroxidase activity of COX-2 enzyme. COX-2 activity was calculated with reference to the total protein in a sample. An unpaired t-test was performed to determine the significance values between the control and treatment groups.
. q-RT PC
Heart tissues were collected from the isolated embryos of both control and treatment groups. Total RNA was isolated from heart tissue using the TRIzol method and quantified through Qubit assay on a Qubit 3.0 fluorimeter (Life Technologies, USA). One µg of isolated RNA was used to prepare cDNA using a high-capacity reverse transcription kit (Applied Biosystems, Thermo Fisher Scientific, USA). Quantitative RT-PCR was performed using LightCycler96 (Roche Diagnostics, USA) as per the following protocol: 32 cycles consisting of denaturation at 95 °C for 10 sec., annealing at 60 °C for 30 seconds, and extension at 72 °C for 30 seconds. The specific products were confirmed by analyzing their melting curves.
Cq values were used for fold change calculation and represented as 2-ΔΔCq according to Livak and Schmittgen (Livak and Schmittgen, 2001). The internal control was 18s rRNA for this experiment. Primers were designed using the online primer blast tool from NCBI (Table 1).
Table 1
Primer sequence
| Gene | Forward primer | Reverse primer | Accession number |
| WNT11 | GACCTGGGTATCGATGGGGA | GGCTTTCAAGACCTGTCTCC | NM_204784 |
| GATA4 | GGCATGCCAACATCGAATTTTT | CTTGTCCGGGGTACTGTGAG | NM_001293106 |
| GATA5 | GGCAAAACCTCAACAGGGTC | CTTGTCCGGGGTACTGTGAG | NM_205421 |
| GATA6 | GCGCCCTACGACGGATCTC | GGGTGGTGGGCACGTAGAC | NM_205420 |
| TBX20 | GATATGCCTACCACCGCTCC | CGTAGAGCCTTGCTGGGAGA | NM_204144 |
| MYOCD | GTCCCCCAACAGCCACTATC | CGTAGAGCCTTGCTGGGAGA | NM_001080715 |
| HAND2 | AGCGGCGATGAGTCTTGTG | CAGCCGTGAAGTAGGGGTTC | NM_204966 |
| SHH | TGCTAGGGATCGGTGGATAG | ACAAGTCAGCCCAGAGGAGA | NM_204821 |
| BMP4 | TGGAAGAACGTGTCCATCGC | AGACTGGCATGGTGGCTCTC | NM_205237 |
| PCNA | TGTTCCTCTCGTTGTGGAGT | TCCCAGTGCAGTTAAGAGCC | NM_204170 |
| CASP3 | AAAGATGGACCACGCTCAGG | TGACAGTCCGGTATCTCGGT | NM_204725 |
| 18s rRNA | GGCCGTTCTTAGTTGGTGGA | GGCCGTTCTTAGTTGGTGGA | XR_005840267 |
In order to minimize biological variation, tissue samples collected from three embryos were pooled, and each variable was analyzed by q-RT PCR, three technical replicates were performed to reduce experimental errors.
Protein expression analysis
Western blot was performed to determine relative protein expression in various groups. Heart tissue was collected from control and treated embryos on days two and three. The tissue was homogenized in lysis buffer with protease inhibitor. Total protein was isolated and quantified by Bradfor’s assay (Bradford, 1976). 30 µg of total protein was electroporated on a 12% polyacrylamide gel and transferred to PVDF membrane, which was then immunostained for MYOCD (Anti-MYOCD IgG raised in mouse, Sigma Aldrich, USA, SAB4200539), PCNA (Anti-PCNA IgG raised in rabbit, Sigma Aldrich, USA, SAB2108448) and Cl.Caspase3 (Anti-Cl.Caspase3 IgG raised in rabbit, Sigma Aldrich, USA, C8487) using primary antibodies diluted 1:1000 times in assay buffer. GAPDH (Anti-GAPDH IgG raised in mouse, Sigma Aldrich, USA) was used as an internal control. Biotinylated secondary polyclonal antibody IgG (Sigma Aldrich, USA, B8895) type was used and probed using ALP-BCIP-NBT for color development of the blots. The blots’ band intensity was measured by densitometry analysis using ImageJ (Supplementary data).
Histological study
Day three control and etoricoxib-treated embryos were isolated using the filter ring method, then rinsed in PBS and fixed in 10% neutral buffer formalin. The tissue was further processed for paraffin wax block preparation. Transverse sections of the heart region were taken using a microtome. These sections were subsequently stained with Harris Hematoxylin and Eosin (Thermo Fisher Scientific, USA). The histological details of the tissue section were visualized using a Leica DM2500 microscope, and pictures were captured using an EC3 camera (utilizing LAS EZ software).
Supplementary Material
Acknowledgements
The work was supported by Innovation in Science Pursuit for Inspired Research (INSPIRE) grant (No. IF190305 Dt. 01/12/2020). BP is thankful to SHODH and DST-INSPIRE for providing fellowship. We are grateful to Sun Pharmaceutical Industries Ltd., Vadodara, India, for providing technical grade Etoricoxib (batch no.7090/F/727/36A) as a generous gift. BP is thankful to Neelanjana and Shivangi for confocal imaging at CIFF, NCBS, Bangalore. We are grateful to Prof. Raj Ladher for providing technical advice, and lab space in NCBS, Banglore.
Abbreviations
COX, Cyclooxygenase ; BMP, Bone morphogenetic protein ; HAND, Heart and Neural Crest Derivatives Expressed ; PGE2, Prostaglandin E2 ; TGF-β, Transforming growth factor-β ; HH, Hamburger and Hamilton stages ; MyoCD, Myocardin ; DAPI, 4',6-diamidino-2-phenylindole ; PCNA, Proliferating cell nuclear antigen ; GAPDH, glyceraldehyde-3-phosphate dehydrogenase ; SHH, sonic hedgehog ; TBX, T-Box Transcription Factor ; ANF, Atrial natriuretic factor ;Declarations
Competing interests
The authors declare that no competing interests exist.
Author contributions
Bhaval Parmar: Investigation, Data curation, Writing – original draft. Urja Verma: Data curation. Juhi Vaishnav: Data curation. Suresh Balakrishnan: Conceptualization, Methodology, Writing – review & editing.
References
Bartelings M. M., Gittenberger-de Groot A. C. (1989). The outflow tract of the heart — embryologic and morphologic correlations. International Journal of Cardiology 22: 289-300.
Bondesen B. A., Mills S. T., Pavlath G. K. (2006). The COX-2 pathway regulates growth of atrophied muscle via multiple mechanisms. American Journal of Physiology-Cell Physiology 290: C1651-C1659.
Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72: 248-254.
Brand T. (2003). Heart development: molecular insights into cardiac specification and early morphogenesis. Developmental Biology 258: 1-19.
Brown D. D., Martz S. N., Binder O., Goetz S. C., Price B. M. J., Smith J. C., Conlon F. L. (2005). Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development 132: 553-563.
Buch P. R., Sarkate P., Uggini G. K., Desai I., Balakrishnan S. (2017). Inhibition of Cyclooxygenase-2 Alters Wnt/β-Catenin Signaling in the Regenerating Tail of Lizard Hemidactylus flaviviridis. Tissue Engineering and Regenerative Medicine 14: 171-178.
Chen J.F., Wang S., Wu Q., Cao D., Nguyen T., Chen Y., Wang D.Z. (2008). Myocardin Marks the Earliest Cardiac Gene Expression and Plays an Important Role in Heart Development. The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology 291: 1200-1211.
Chen J. (2000). Genetics of heart development. Trends in Genetics 16: 383-388.
Chen J.N., van Eeden F.J., Warren K.S., Chin A., Nusslein-Volhard C., Haffter P., Fishman M.C. (1997). Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124: 4373-4382.
Dees E., Lin H., Cotton R. B., Graham T. P., Dodd D. A. (2000). Outcome of preterm infants with congenital heart disease. The Journal of Pediatrics 137: 653-659.
Dyer L. A., Kirby M. L. (2009). Sonic hedgehog maintains proliferation in secondary heart field progenitors and is required for normal arterial pole formation. Developmental Biology 330: 305-317.
Eisenberg C. A., Eisenberg L. M. (1999). WNT11 promotes cardiac tissue formation of early mesoderm. Developmental Dynamics 216: 45-58.
Hamburger V., Hamilton H. L. (1951). A series of normal stages in the development of the chick embryo. Journal of Morphology 88: 49-92.
Huang J., Min Lu M., Cheng L., Yuan L.J., Zhu X., Stout A. L., Chen M., Li J., Parmacek M. S. (2009). Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proceedings of the National Academy of Sciences 106: 18734-18739.
Jana S., Chatterjee K., Ray A. K., DasMahapatra P., Swarnakar S. (2016). Regulation of Matrix Metalloproteinase-2 Activity by COX-2-PGE2-pAKT Axis Promotes Angiogenesis in Endometriosis. PLOS ONE 11: e0163540.
Khoshnood B., Lelong N., Houyel L., Thieulin A.C., Jouannic J.M., Magnier S., Delezoide A.L., Magny J.F., Rambaud C., Bonnet D., Goffinet F. (2012). Prevalence, timing of diagnosis and mortality of newborns with congenital heart defects: a population-based study. Heart 98: 1667-1673.
Laforest B., Andelfinger G., Nemer M. (2011). Loss of Gata5 in mice leads to bicuspid aortic valve. Journal of Clinical Investigation 121: 2876-2887.
Laforest B., Nemer M. (2011). GATA5 interacts with GATA4 and GATA6 in outflow tract development. Developmental Biology 358: 368-378.
Laurent F., Girdziusaite A., Gamart J., Barozzi I., Osterwalder M., Akiyama J. A., Lincoln J., Lopez-Rios J., Visel A., Zuniga A., Zeller R. (2017). HAND2 Target Gene Regulatory Networks Control Atrioventricular Canal and Cardiac Valve Development. Cell Reports 19: 1602-1613.
Lepore J. J. (2006). GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. Journal of Clinical Investigation 116: 929-939.
Linask K. K., Han M., Cai D. H., Brauer P. R., Maisastry S. M. (2005). Cardiac morphogenesis: Matrix metalloproteinase coordination of cellular mechanisms underlying heart tube formation and directionality of looping. Developmental Dynamics 233: 739-753.
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402-408.
Lopez-Sanchez C., Garcia-Masa N., Ganan C. M., Garcia-Martinez V. (2009). Movement and commitment of primitive streak precardiac cells during cardiogenesis. The International Journal of Developmental Biology 53: 1445-1455.
Lutin W.A., Brumund M.R., Jones C., Tharpe C.E., Montegomery M., McCaffrey F.M. (1999). Hemodynamic Abnormalities in Fetuses with Congenital Heart Disease. Pediatric Cardiology 20: 390-395.
Madonna R., Taylor D. A., Geng Y.J., De Caterina R., Shelat H., Perin E. C., Willerson J. T. (2013). Transplantation of Mesenchymal Cells Rejuvenated by the Overexpression of Telomerase and Myocardin Promotes Revascularization and Tissue Repair in a Murine Model of Hindlimb Ischemia. Circulation Research 113: 902-914.
Molkentin J. D., Lin Q., Duncan S. A., Olson E. N. (1997). Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis.. Genes & Development 11: 1061-1072.
Narita N., Bielinska M., Wilson D.B. (1997). Cardiomyocyte differentiation by GATA-4-deficient embryonic stem cells. Development 124: 3755-3764.
Neil J. R., Johnson K. M., Nemenoff R. A., Schiemann W. P. (2008). Cox-2 inactivates Smad signaling and enhances EMT stimulated by TGF- through a PGE2-dependent mechanisms. Carcinogenesis 29: 2227-2235.
Nemer M. (2008). Genetic insights into normal and abnormal heart development. Cardiovascular Pathology 17: 48-54.
Nørregaard R., Kwon T.H., Frøkiær J. (2015). Physiology and pathophysiology of cyclooxygenase-2 and prostaglandin E2 in the kidney. Kidney Research and Clinical Practice 34: 194-200.
Olson E. N. (2006). Gene Regulatory Networks in the Evolution and Development of the Heart. Science 313: 1922-1927.
Pakrasi P. L., Jain A. K. (2008). Cyclooxygenase-2-derived endogenous prostacyclin reduces apoptosis and enhances embryo viability in mouse. Prostaglandins, Leukotrienes and Essential Fatty Acids 79: 27-33.
Pandur P., Läsche M., Eisenberg L. M., Kühl M. (2002). Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature 418: 636-641.
Pérez-Pomares J. M., de la Pompa J. L. (2011). Signaling During Epicardium and Coronary Vessel Development. Circulation Research 109: 1429-1442.
Peterkin T., Gibson A., Patient R. (2007). Redundancy and evolution of GATA factor requirements in development of the myocardium. Developmental Biology 311: 623-635.
Piven O. O., Winata C. L. (2017). The canonical way to make a heart: β-catenin and plakoglobin in heart development and remodeling. Experimental Biology and Medicine 242: 1735-1745.
Plageman T. F., Yutzey K. E. (2004). Differential Expression and Function of Tbx5 and Tbx20 in Cardiac Development. Journal of Biological Chemistry 279: 19026-19034.
Randall C. L., Anton R. F., Becker H. C., Hale R. L., Ekblad U. (1991). Aspirin dose-dependently reduces alcohol-induced birth defects and prostaglandin E levels in mice. Teratology 44: 521-529.
Raphel L., Talasila A., Cheung C., Sinha S. (2012). Myocardin Overexpression Is Sufficient for Promoting the Development of a Mature Smooth Muscle Cell-Like Phenotype from Human Embryonic Stem Cells. PLoS ONE 7: e44052.
Ricciotti E., FitzGerald G. A. (2011). Prostaglandins and Inflammation. Arteriosclerosis, Thrombosis, and Vascular Biology 31: 986-1000.
Shim M., Foley J., Anna C., Mishina Y., Eling T. (2010). Embryonic Expression of Cyclooxygenase-2 Causes Malformations in Axial Skeleton. Journal of Biological Chemistry 285: 16206-16217.
Singh M. K., Li Y., Li S., Cobb R. M., Zhou D., Lu M. M., Epstein J. A., Morrisey E. E., Gruber P. J. (2010). Gata4 and Gata5 Cooperatively Regulate Cardiac Myocyte Proliferation in Mice. Journal of Biological Chemistry 285: 1765-1772.
Srivastava D., Thomas T., Lin Q., Kirby M. L., Brown D., Olson E. N. (1997). Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nature Genetics 16: 154-160.
Stennard F. A., Costa M. W., Elliott D. A., Rankin S., Haast S. J.P., Lai D., McDonald L. P.A., Niederreither K., Dolle P., Bruneau B. G., Zorn A. M., Harvey R. P. (2003). Cardiac T-box factor Tbx20 directly interacts with Nkx2-5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Developmental Biology 262: 206-224.
Tsuchihashi T., Maeda J., Shin C. H., Ivey K. N., Black B. L., Olson E. N., Yamagishi H., Srivastava D. (2011). Hand2 function in second heart field progenitors is essential for cardiogenesis. Developmental Biology 351: 62-69.
Tsuda T., Majumder K., Linask K. K. (1998). Differential expression of flectin in the extracellular matrix and left-right asymmetry in mouse embryonic heart during looping stages. Developmental Genetics 23: 203-214.
van Berlo J. H., Maillet M., Molkentin J. D. (2013). Signaling effectors underlying pathologic growth and remodeling of the heart. Journal of Clinical Investigation 123: 37-45.
van den Hoff M. J.B., Moorman A. F.M., Ruijter J. M., Lamers W. H., Bennington R. W., Markwald R. R., Wessels A. (1999). Myocardialization of the Cardiac Outflow Tract. Developmental Biology 212: 477-490.
Verma U., Gautam M., Parmar B., Khaire K., Wishart D. S., Balakrishnan S. (2021). New insights into the obligatory nature of cyclooxygenase-2 and PGE2 during early chick embryogenesis. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1866: 158889.
Xu D., Bu J., Gu S., Xia Y., Du J., Wang Y. (2011). Celecoxib Impairs Heart Development via Inhibiting Cyclooxygenase-2 Activity in Zebrafish Embryos . Anesthesiology 114: 391-400.
Ye B., Li L., Xu H., Chen Y., Li F. (2019). Opposing roles of TCF7/LEF1 and TCF7L2 in cyclin D2 and Bmp4 expression and cardiomyocyte cell cycle control during late heart development. Laboratory Investigation 99: 807-818.
Yelbuz T. M., Waldo K. L., Kumiski D. H., Stadt H. A., Wolfe R. R., Leatherbury L., Kirby M. L. (2002). Shortened Outflow Tract Leads to Altered Cardiac Looping After Neural Crest Ablation. Circulation 106: 504-510.
Zhao Z., Rivkees S. A. (2000). Programmed cell death in the developing heart: Regulation by BMP4 and FGF2. Developmental Dynamics 217: 388-400.