Int. J. Dev. Biol. 67: 1 - 8 (2023)
DNA methyltransferase (Dnmt) silencing causes increased Cdx2 and Nanog levels in surviving embryos
Open Access | Original Article | Published: 4 May 2023
Abstract
Epigenetic mechanisms are one of the essential regulators of gene expression which do not involve altering the primary nucleotide sequence. DNA methylation is considered among the most prominent epigenetic mechanisms in controlling the functions of genes related to cell differentiation, cell cycle, cell survival, autophagy, and embryo development. DNA methyl transferases (Dnmts) control DNA methylation, the levels of which are differentially altered during embryonic development, and may determine cell differentiation fate as in the case of pluripotent inner cell mass (ICM) or trophectoderm (TE). In this study, we aimed to analyze the role of Dnmt1 and Dnmt3a enzymes in ICM (using the Nanog marker) and TE (using the Cdx2 marker) differentiation, autophagy (using p62 marker), reactive oxygen species (ROS) production, and apoptosis (using TUNEL) during mouse preimplantation embryo development. Following knockdown of Dnmt1 and Dnmt3a in zygotes, expression levels of Cdx2 in the trophectoderm and Nanog in the inner cell mass were measured, as well as p62 levels, reactive oxygen species (ROS) production, and apoptosis levels after 96 hours in embryo culture. We found that knockdown of Dnmt1 or Dnmt3a significantly induced Cdx2 and Nanog expression. Similarly, p62 expression, ROS levels and apoptosis significantly increased after silencing. This study shows that Dnmt genes are highly crucial for embryonic fate determination and survival. Further studies are required to reveal the specific targets of these methylation processes related to cell differentiation, survival, autophagy, and ROS production in mouse and human preimplantation embryos.
Keywords
DNA methylation, Dnmt, cell differentiation, preimplantation embryo development, siRNA
Introduction
Formation of a blastocyst shows a unique differentiation from totipotent embryo to pluripotent inner cell mass (ICM) and trophectoderm (TE) in mammals (Watson and Barcroft 2001; Adjaye et al., 2005). While the TE constitutes the extra-embryonic membranes such as the yolk sac, amniotic membrane, and placenta, the ICM gives rise to embryonic tissues (Rossant 2004; Yamanaka et al., 2006). It is known that this differentiation is prone the exogenous effects and in vitro culture conditions during preimplantation embryo development may harm first-cell differentiation (Niemann and Wrenzycki 2000; Schultz and Williams 2002; Gao et al., 2003). In this process, many mechanisms and pathways are responsible for the formation of a blastocyst, which later triggers a series of cellular and molecular events for further differentiation (Zhang Y et al., 2007). By ensuring the activation and repression of developmental-related genes, epigenetic mechanisms consist of two processes: histone modifications and DNA methylation (Bannister and Kouzarides 2011; Cao and Yan 2012; Du et al., 2015). DNA methylation is carried out by adding a methyl group to the fifth carbon atom of the cytosine residues using S-adenosyl-L-methionine (AdoMet) as a methyl donor. Two different DNA methylation processes, de novo, and maintenance methylation, which are under the control of DNA methyltransferases (Dnmts) (Turek-Plewa and Jagodzinski 2005). Dnmt1 mainly plays role in the maintenance of hemi-methylated strands during DNA replication, whereas Dnmt3a is responsible for de novo methylation (Fatemi et al., 2002).
The caudal-type homeodomain protein Cdx2 is implicated in the formation and maintenance of the TE (Sritanaudomchai et al., 2009), NANOG determines inner cell mass fate (Harvey et al., 2009). DNA methylation plays a key role in the first cell-lineage specification of blastocysts into TE and ICM. However, it remains unclear as to which of the DNA methyltransferase(s) acts on this process. Assou et al., (2012) described the localization of the Dnmt3a and Dnmt1 expression in human TE cells (Assou et al., 2012), Galán et al., (2010) reported that Dnmt3a is highly expressed in ICM cells (Galan et al., 2010); but Petrussa et al., (2014) found no difference between two cell population (Petrussa et al., 2014). It can be postulated that those prominent differences may derive from the obtained blastocyst by using different in vitro culture conditions. Studies based on experimental animal models have found that DNA methylation levels can change depending on the varying in vitro culture conditions (Uysal et al., 2015).
Autophagy is a biological process in which macromolecules or damaged organelles are broken down by the lysosome (He et al., 2023). Research on the maintenance of reproductive capacity highlights the contribution of autophagy in maintaining primordial follicle numbers, regulating the pregnancy process with the involvement of hormone synthesis, and improving age-related declining ovarian reserve (Gawriluk et al., 2014; Song et al., 2015; Huang et al., 2022). Recent studies show that p62, a selective autophagy adapter protein for lysosomal degradation, can recognize the glycolytic enzymes crucial for the selective degradation process and is essential for maintaining female fertility (Li et al., 2021; He et al., 2023). Autophagy is also critical for the fate of blastomeres as it degrades Nanog via Atg7-dependent autophagy pathway (Zhou et al., 2022). Atg7-dependent autophagic pathway activity depends on the multifunctional adaptor protein p62 (Kim et al., 2019). On the other hand, it has been demonstrated that the epigenome of the p62 promoter is collectively regulated by Dnmt1 and HDAC1 (Lee et al., 2021). However, the relationship between Dnmt enzymes and p62 activation in embryo development remains elusive. To test, if knockdown of Dnmt1 or Dnmt3a enzymes, and therefore increased degeneration rates, can be related to the autophagic process, we analyzed p62, which is considered a player of initiating autophagosome membrane formation around protein aggregates by promoting recruitment and assembly of upstream Atg proteins to the initiation sites.
Oxidative stress refers to the imbalance in the redox system when free radicals increase to a level that exceeds the scavenging capacity of the endogenous antioxidant system. Ninety-five percent of free radicals belong to reactive oxygen species (ROS) (Wang et al., 2021). ROS is a double-edged sword for oocyte and embryo development. Normally, ROS are essential in mediating folliculogenesis, meiosis, ovulation and embryonic development as secondary messengers for cellular signaling (Agarwal et al., 2012). However, when free radicals are overproduced, intracellular ROS can accumulate and then attack biological macromolecules and organelles, leading to DNA damage and apoptosis (Roth 2018).
During preimplantation development, global DNA methylation is gradually erased, starting from the zygote by the demethylation process, and then established by the de novo methylation mechanism around the blastocyst stage (Saitou et al., 2012). Previously, we have comprehensively evaluated the spatial-temporal distribution and expression levels of the Dnmt1 and Dnmt3a proteins in mouse preimplantation embryos (Uysal et al., 2017). We clearly documented the stage-dependent alteration of the levels of these proteins. Their levels were high in zygotes, then gradually decreased until morula stage during embryo development and ultimately, re-increased in the blastocyst stage, showing a parallel pattern with the global DNA methylation levels (Uysal et al., 2017). In another study, we also revealed the crucial roles of DNA methylation and Dnmts expression in preimplantation embryo development. We found that silencing of Dnmt1 or Dnmt3a genes resulted to embryo development arrests and blastomere degeneration during preimplantation embryo development (Uysal et al., 2021). On the other hand, survived embryos caused to raise new questions about how they did so. Therefore, the current study was designed to respond to some of the questions that what the effects of DNA methylation via either Dnmt1 or Dnmt3a on cell lineage differentiation, apoptosis and ROS levels are. Briefly, we found that knocking down of Dnmt1 or Dnmt3a leads to increase both Cdx2 and Nanog levels in survived blastocysts. We also found that p62 expression, reactive oxygen species (ROS) levels and apoptosis significantly increased after silencing. Our results suggested that Dnmt proteins are critical and may contribute to cell lineage differentiation and survival pathways in preimplantation embryo development.
Results
Cdx2 expression level
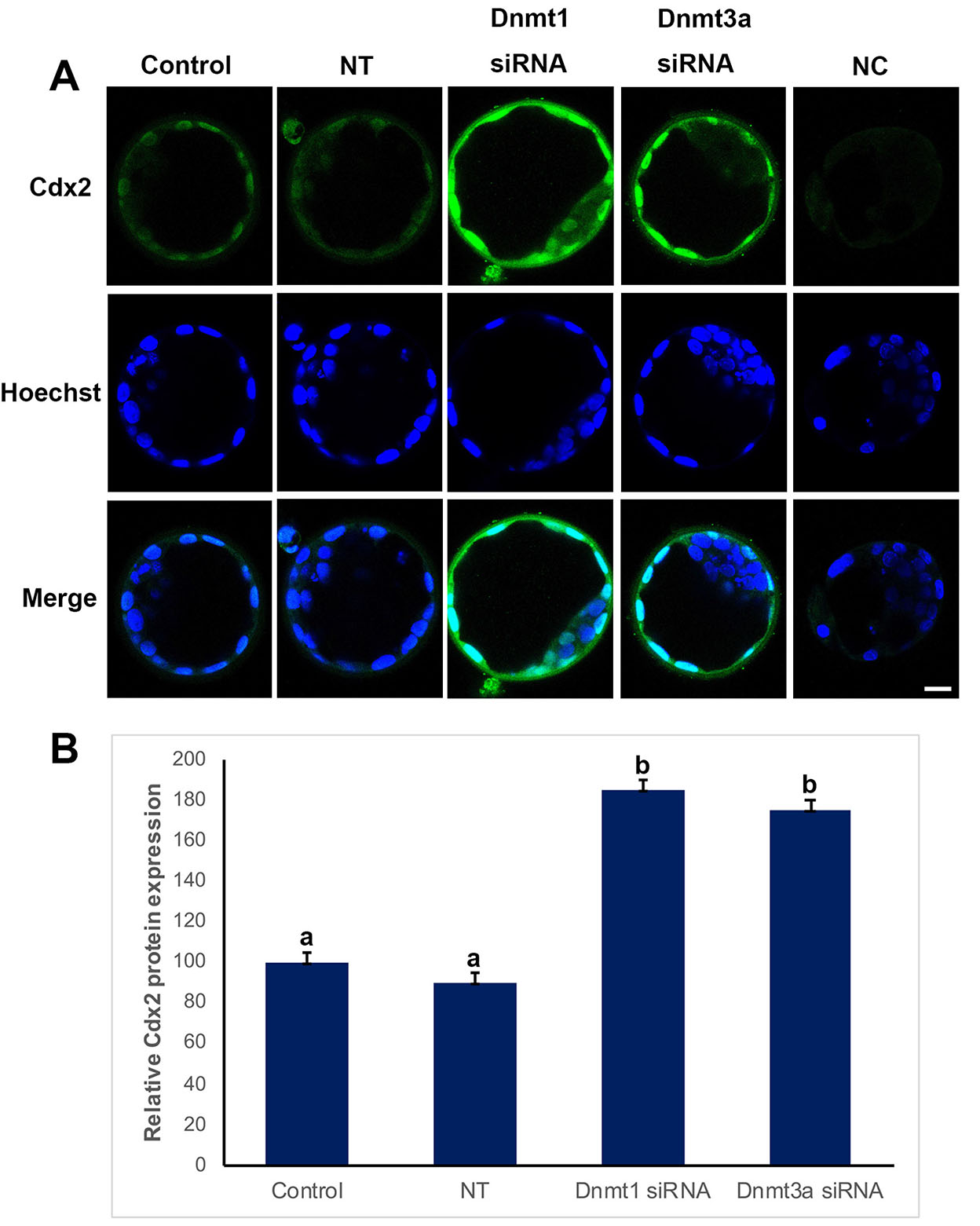
To analyze, the effects of Dnmt1 or Dnmt3a silencing on trophectoderm layer, we examined Cdx2 protein expression (Fig. 1). Cdx2 signal was found in the nuclei of trophoblast cells. The expression level of control group was considered as 100% and the other groups were evaluated based on this value. The non-target (90%) group was similar to that of the control, while Cdx2 levels significantly increased in Dnmt1 or Dnmt3a silenced groups as 185% and 175%, respectively (p<0.05) (Fig. 1B).
Fig. 1. Cdx2 expression in control, non-targeted (NT), Dnmt1- and Dnmt3a-siRNA-treated blastocysts.
(A) Immunofluorescence (IF) assays obtained from super-resolution 3D image reconstructions of Cdx2 signals. Cdx2 expression significantly increased in siRNA groups. (B) Relative Cdx2 expression levels in IF analysis. The statistical significance was determined by using one-way ANOVA followed by Dunn’s post hoc test. P<0.05 was considered statistically significant, as indicated by different letters on the columns. Bars in graphs represent mean ± SD. Scale bar: 20 µm; NC: Negative control.
Nanog expression level
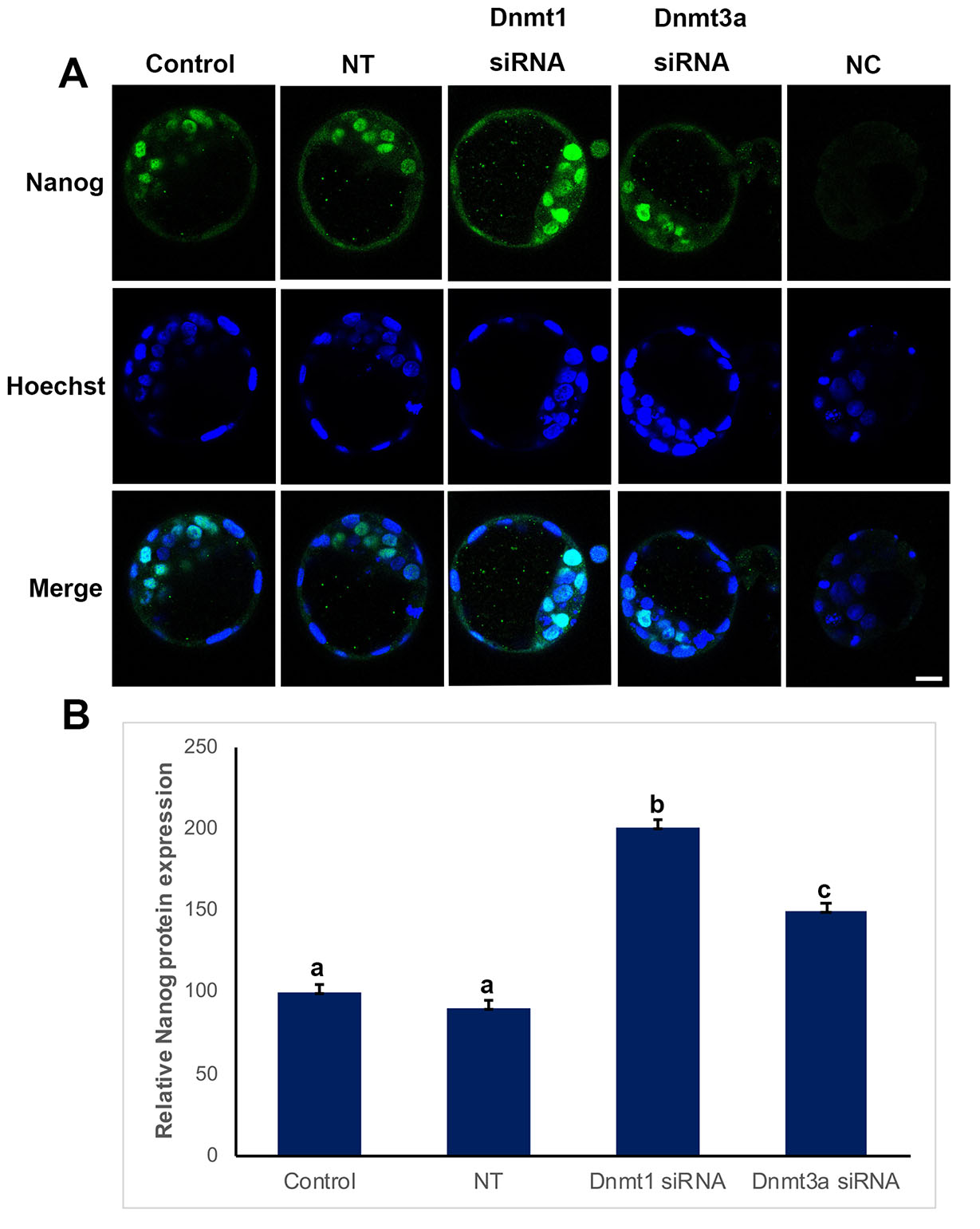
Nanog protein expression levels were analyzed to evaluate the effect of Dnmt1 or Dnmt3a silencing on inner cell mass (Fig. 2). Nanog expression level of control (100%) and non-target (90%) groups showed no statistical significance, while it significantly increased when Dnmt1 or Dnmt3a were silenced as 200% and 150%, respectively (p<0.05) (Fig. 2B).
Fig. 2. Nanog expression in control, non-targeted (NT), Dnmt1- and Dnmt3a-siRNA-treated blastocysts.
(A) Immunofluorescence (IF) assays obtained from super-resolution 3D image reconstructions of Nanog signals. Nanog expression significantly increased in siRNA groups. (B) Relative Nanog expression levels in IF analysis. The statistical significance was determined by using one-way ANOVA followed by Dunn’s post hoc test. P<0.05 was considered statistically significant, as indicated by different letters on the columns. Bars in graphs represent mean ± SD. Scale bar: 20 µm; NC: Negative control.
Cell death rates
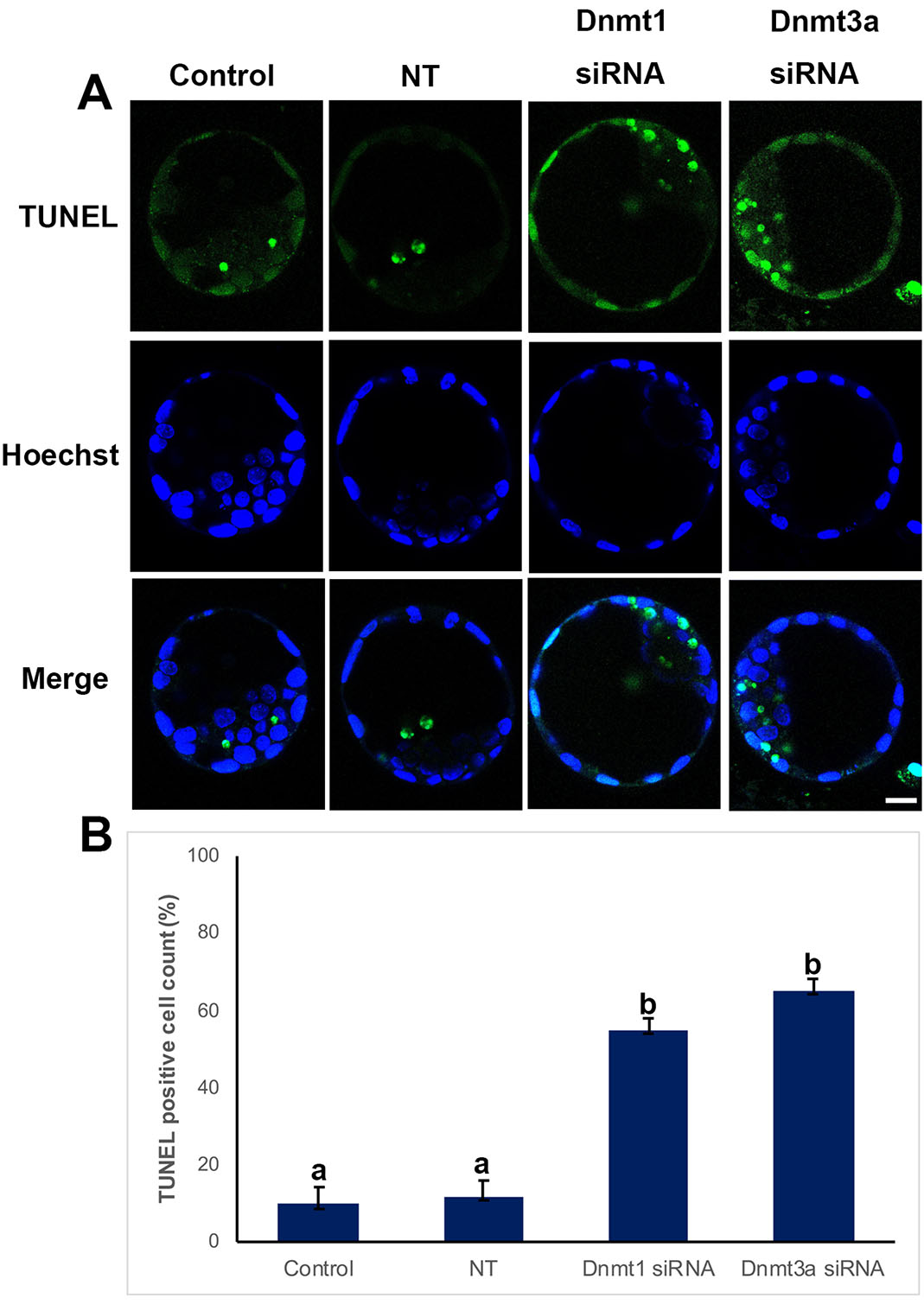
When we analyzed apoptotic process, we found that control and non-target groups revealed no statistical significance (Fig. 3). Interestingly, there were significantly increased apoptotic cells in Dnmt1 and Dnmt3a siRNA groups (p<0.05) (Fig. 3B).
Fig. 3. TUNEL assay in control, non-targeted (NT), and siRNA-treated blastocysts.
(A) Immunofluorescence (IF) assays obtained from super-resolution 3D image reconstructions of apoptotic cells. Apoptotic cell levels significantly increased in the siRNA group. (B) TUNEL positive cells in IF analysis. Statistical significance was determined by using one-way ANOVA followed by Dunn’s post hoc test. P<0.05 was considered statistically significant, as indicated by different letters on the columns. Bars in graphs represent mean ± SD. Scale bar: 20 µm.
P62 expression level
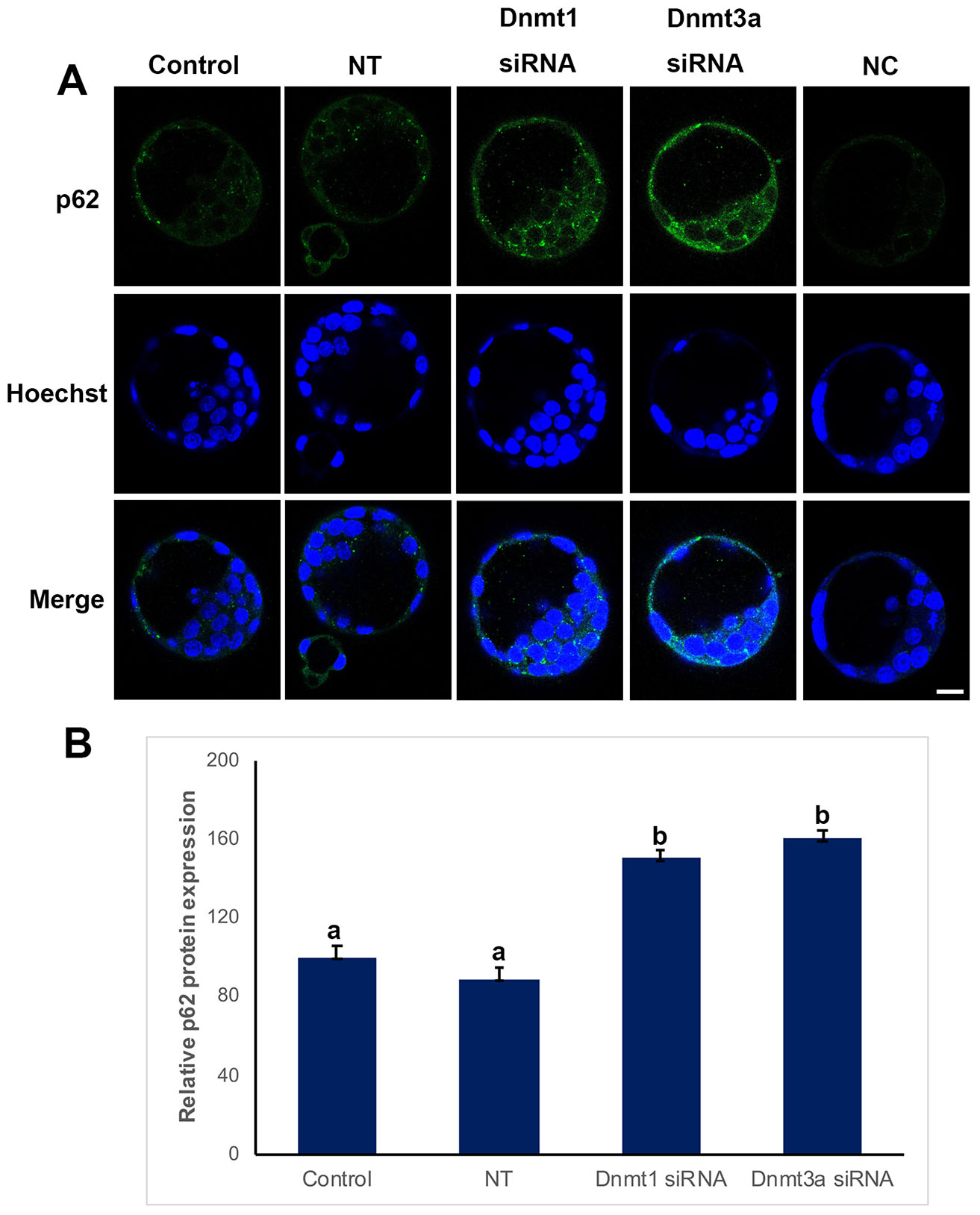
Immunofluorescent analysis of p62 protein showed that p62 is localized in cytoplasm, no signal was observed in cell nucleus (Fig. 4). When the levels of p62 expression were analyzed, control (100%) and non-target (89%) groups were comparable, while a significant increase was observed in siRNA groups, 150% for Dnmt1 siRNA, 160% for Dnmt3a siRNA (p<0.05) (Fig. 4B).
Fig. 4. p62 expression in control, non-targeted (NT), and siRNA-treated blastocysts.
(A) Immunofluorescence (IF) assays obtained from super-resolution 3D image reconstructions of p62-signals. P62 expression significantly increased in siRNA groups. (B) Relative P62 protein expression levels in IF analysis. Statistical significance was determined by using one-way ANOVA followed by Dunn’s post hoc test. P<0.05 was considered statistically significant, as indicated by different letters on the columns. Bars in graphs represent mean ± SD. Scale bar: 20 µm. NC: Negative control.
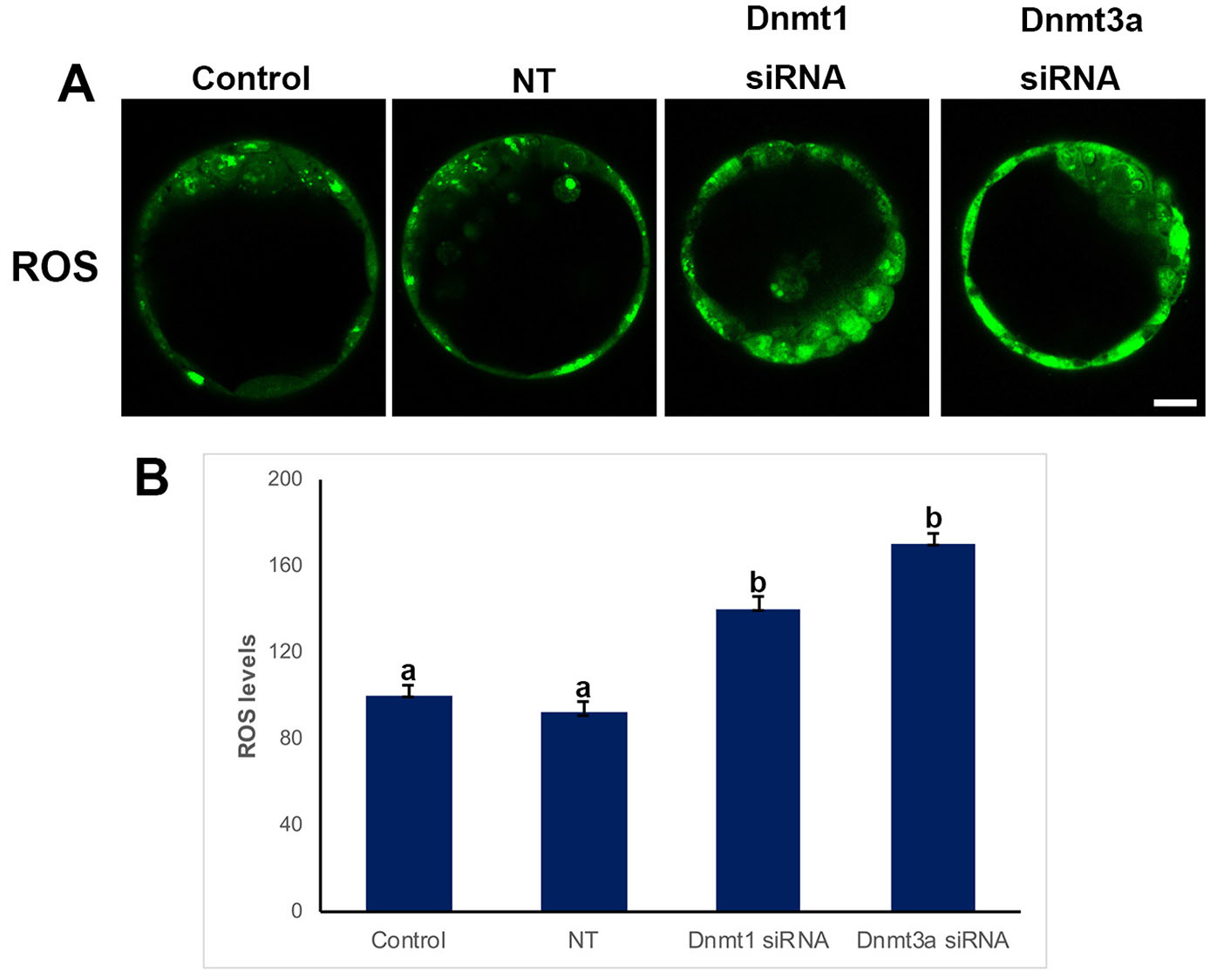
ROS levels
We performed ROS marking to investigate whether ROS levels were affected by Dnmt silencing (Fig. 5). We found that ROS levels were significantly increased when Dnmt1 or Dnmt3a were silenced, 140% and 170%, respectively (p<0.05). ROS levels did not show any difference between the non-target (92%) and control groups (100%) (Fig. 5B).
Fig. 5. Reactive oxygen species (ROS) levels in control, non-targeted (NT), and siRNA-treated blastocysts.
(A) Immunofluorescence (IF) assays obtained from super-resolution 3D image reconstructions of ROS signals. ROS levels significantly increased in siRNA groups. (B) Relative ROS levels in IF analysis. Statistical significance was determined by using one-way ANOVA followed by Dunn’s post hoc test. P<0.05 was considered statistically significant, as indicated by different letters on the columns. Bars in graphs represent mean ± SD. Scale bar: 20 µm.
Discussion
Mammalian preimplantation embryo development involves fundamental and successive events, such as embryonic genome activation, compaction, cavitation, polarization, cleavage divisions, and lineage segregation (Sozen et al., 2014). In this process, many mechanisms and pathways are responsible for proper embryo development, which triggers a series of cellular and molecular events. By ensuring the activation and repression of developmental related genes, epigenetic mechanisms consist of two process (i) histone modifications and (ii) DNA methylation. Following fertilization, the expression of Dnmt genes gradually decreases from zygote to 16-cell/morula stage embryos and thus the global DNA methylation level and consequently the paternally and maternally imprinted genes are demethylated thereafter (Saitou et al., 2012). Recently, we published that preimplantation embryo development is severely impaired from Dnmt1 or Dnmt3a silencing (Uysal et al., 2021). Among the other epigenetic mechanisms, DNA methylation can be considered as the most prominent in controlling the functions of genes including cell cycle-related ones. DNA methylation is involved in the regulation of the expression of genes during oogenesis, spermatogenesis, and embryo development. Although it is known that abnormal gene expressions may pause the embryo development process, it has not been clarified which mechanism controls these genes. In the present study, we aimed to reveal the effects of Dnmt1 and Dnmt3a in first cell lineage differentiation, autophagy, cell death and production of ROS by knocking down of either Dnmt1 or Dnmt3a genes with siRNA during preimplantation embryo development from zygote to blastocysts.
We found that Cdx2 and Nanog expression significantly increased when Dnmt1 or Dnmt3a was silenced. Nanog as a pluripotent cell-specific gene plays important roles in regulation of signaling pathways for maintenance and induction of pluripotency in inner cell mass and embryonic stem cells in mouse (Habibi et al., 2018), and progressively decreases during embryonic stem cells differentiation (Grubelnik et al., 2020). It was recently shown that SQSTM1/p62 could translocate into the nucleus and directly bind with K63-polyubiquitinated NANOG for protein degradation (Zhou et al., 2022). Here, we observed increased Nanog expression in nucleus and increased p62 levels in cytoplasm in siRNA groups. Therefore, diminished transportation of p62 from cytoplasm to nucleus may result in increased levels of both p62 and Nanog. Dnmt1 in embryonic stem cells isolated from wild-type blastocysts keeps the trophectodermal transcription factor gene E74-like factor 5 (Elf5) hypermethylated, a state by which trophectoderm-specific transcription factors such as Cdx2 and Eomes are suppressed, therefore further differentiation of trophectoderm is temporarily inhibited (Ng et al., 2008; Smith and Meissner 2013). It was demonstrated that CDX2 mRNA was not expressed in embryos through the 1-cell to 4-cell stages in mouse embryos, and the expression gradually increased from 8-cell to blastocyst stages. Moreover, CDX2 siRNA injection into mouse fertilized eggs resulted in development blockage at 8–10-cell stage (Zhang J et al., 2006), and re-expression of CDX2 could rescue developmental block defects (Zhang W et al., 2018). Collectively, these findings suggest that CDX2 may play an important role in the processes of early embryonic development. Although Dnmt enzymes, thus the methylation level, gradually decreased from zygote to 16-cell/morula stage embryos, then re-increased in blastocysts. This finding clearly indicates the importance of timely regulated methylation events. In blastocysts, the timing of methylation/demethylation event is very critical in both trophectoderm and inner cell mass. When Dnmt1 or Dnmt3a is knocked-down in a zygote, in parallel with the severe alteration in global methylation, the suppressive effects of methylation in blastomere-specific genes and transcription factors would be deteriorated. Some embryos would be able to get rid of these destructive effects of altered methylation of certain genes due to knockdown of Dnmts via various alterative mechanisms including autophagy related ones.
Dnmts are crucial to control the activity of cell cycle-related proteins. It was shown that Dnmt inhibitors led to cell cycle arrest (Xiong et al., 2009), cell death (Cui et al., 2010), and cell differentiation (Vivaldi et al., 2009) in various cancer cells. We previously analyzed embryo development rates after knocking down of Dnmt1 or Dnmt3a and found a significant decrease to reach blastocyst stage. This finding suggests that these enzymes are also essential for early embryo development. In the current study, we aimed to analyze the effect of Dnmt1 and Dnmt3a on cell death in survived embryos to elucidate if survived embryos are composed of healthy cells. Results revealed that inhibition of Dnmt1 or Dnmt3a caused increased cell death even if embryo development continue, and this finding indicates that cell survival mechanism is negatively affected by the diminished levels of Dnmts. ROS can be one of the explanations for increased cell death as Dnmt silencing increased the ROS levels. It was shown that a decrease in Dnmt levels induces ROS levels in somatic cells (Kornicka et al., 2017) and the strong relationships between ROS levels and apoptosis and also autophagy were indicated in many studies (Xu et al., 2011; Luo et al., 2019).
One of the classical receptors of autophagy is p62, which was the first selected autophagy adaptor discovered in mammals. p62 is a multifunctional protein located throughout the cell and involved in many signal transduction pathways, also involved in the proteasomal degradation of ubiquitinated proteins (Liu et al., 2016). Here, we showed that a decrease of Dnmt1 or Dnmt3a causes an increase in p62 levels. This finding suggests that Dnmts are vital enzymes to control autophagic mechanism in embryos.
In conclusion, the current study demonstrates that Dnmt1 and Dnmt3a may have various effects on early cell differentiation and cell survival. We show here that some of embryos can survive after Dnmt silencing, thereby tolerating increased ROS levels and apoptotic cell death. We think that an activated autophagic mechanism may contribute to survival, with affected cells striving to remove disrupted components. Further studies such as gene sequencing after knocking down of these genes are needed to clarify which pathways are affected and thus necessary for these processes (Xue et al., 2013; Zhao et al., 2019). Other genes, particularly those related to autophagy, should be analyzed in this process. Although this point could be considered to be a limitation of the present study, our results are nevertheless fundamental to justify additional studies of the possible roles of Dnmts in mouse cell differentiation, as well as in embryo development and implantation.
Materials and Methods
Animals
The experimental protocol was approved by the Animal Care and Usage Committee of Ankara University (Protocol no: 2022-12-101). Female Balb/C mice at 4-6 weeks and male mice at 8-10 weeks of age were purchased from the Research Animal Laboratory Unit. All mice were hosted with free access to food and water and kept in a 12-hr light/dark cycle.
Collection of Zygotes and in vitro Culture
To collect early embryos, mice were injected intraperitoneally with 5 IU PMSG (pregnant mare's serum gonadotropin) (Intervet). Forty-eight hours after PMSG treatment, 5 IU hCG (human chorionic gonadotropin) (Sigma-Aldrich, USA) were injected into the PMSG-primed mice and they were kept with mature male mice at a rate of 1 female:1 male overnight for mating. Next morning, the presence of vaginal plug verified the fertilization. Pregnant female mice were used to obtain zygotes from oviducts at 20 h following hCG injection. Notably, the cumulus cells surrounding the zygotes were removed using hyaluronidase (Vitrolife, Sweden) solution at a concentration of 1 mg/mL. Embryos were immediately placed in morpholinepropanesulfonic acid (MOPS)-buffered medium (G-MOPSTM) (Vitrolife, Sweden) after collection and then transferred to culture medium (G-TLTM; Vitrolife, Sweden) as 50 μL volumes of culture drops in 35 mm culture dishes (Corning, USA) that were overlaid by approximately 3 mL of heavy paraffin oil (Sigma-Aldrich, USA). Zygotes (0 h) were cultured up to the blastocyst stage (96 h) at 37°C in 6% CO2. A total of 330 embryos for control group, 325 for non-targeted group, 310 for Dnmt1-siRNA group, 315 for Dnmt3a-siRNA group and 310 embryos for negative control group were evaluated in at least three replicated experiments.
siRNA Treatment
siRNA treatments were performed according to our previously published manuscript (Uysal et al., 2021). Briefly, zygotes were transfected with Dnmt1 or Dnmt3a-specific small interfering RNA (siRNA) duplexes vs non-targeting control siRNA duplexes at 50 nM for 96 h using DharmaFECT (Dharmacon, USA) as a transfection reagent. Nontargeting siRNA duplex served as the negative control. Zygotes were co-transfected with a 1 nM siGLO green transfection indicator (Dharmacon, USA) to verify the success of transfection. The siGLO green transfection indicator (absorbance/emission max is 494/520 nm, Dharmacon, USA) was used to determine the optimal siRNA transfection conditions.
Immunofluorescence (IF) Staining
Blastocysts were fixed and then permeabilized with 4% paraformaldehyde (Sigma-Aldrich, USA) solution, and 1% Tween-20 (Sigma-Aldrich, USA) at room temperature (RT), respectively. They were blocked with blocking solution including 20% normal goat serum (Vector Laboratory, USA), and then IF were applied to detect the relative quantity and cellular distribution profiles of the Cdx2, Nanog and p62 proteins in the blastocysts. Briefly, blastocysts were incubated overnight at +4°C with rabbit polyclonal antibody against Cdx2 [Abcam, UK; ab76541], rabbit monoclonal antibodies against Nanog (Cell Signaling, 8822) or p62 (Cell Signaling, 23214). After a triple wash with 1x PBS including 2% bovine serum albumin (BSA) for 10 min each (PBS-BSA; Sigma-Aldrich, USA), blastocysts were incubated with anti-rabbit Alexa 488 secondary antibody (Invitrogen, USA) for 1 h at RT followed by triple washes with 1x PBS-BSA solution for 10 min each. The omission of primary antibodies served as controls. All staining steps were performed using mini well trays (Thermo Fisher Scientific, USA) in a humidified chamber. Stained blastocysts were gently transferred onto glass-bottomed 35-mm Petri dishes in a 4 μL-drop of PBS-based mounting medium containing 1 μg/mL Hoechst 33342 (Thermo Fisher Scientific, USA) for DNA labeling. The top was covered with a droplet of paraffin oil (Ovoil, Vitrolife, Sweden). All fluorescently tagged blastocysts were kept intact in terms of their 3D spherical shape and then were examined and imaged using a Zeiss LSM-880 Airyscan® system (Zeiss, Germany). All groups of blastocysts were stained at least five times and analyses were performed in cumulated images. The signal intensities were measured by Image J software (National Institutes of Health, Bethesda, Maryland, USA), and the relative staining intensity levels of the Cdx2, Nanog and P62 proteins have been quantified.
TUNEL assay
TUNEL assay was performed by In situ Cell Death Detection Kit (Sigma-Aldrich, USA) according to the manufacturer’s instructions. Following fixation of blastocysts in 4% paraformaldehyde (Sigma-Aldrich, USA), and then permeabilized with 1% Tween-20 (Sigma-Aldrich, USA) at room temperature (RT), embryos were washed three times in PBS. Fixed embryos were incubated with TUNEL reaction mixture, and for negative control, embryos were incubated with TUNEL label solution (Sigma-Aldrich) only for 1 h at 37°C in the dark. Stained blastocysts were gently transferred onto glass-bottomed 35-mm Petri dishes in a 4 μL-drop of PBS-based mounting medium containing 1 μg/mL Hoechst 33342 (Thermo Fisher Scientific, USA). The top was covered with a droplet of paraffin oil. All fluorescently tagged blastocysts were kept intact in terms of their 3D spherical shape and then were examined and imaged using a Zeiss LSM-880 Airyscan® system (Zeiss, Germany). Total number of nuclei and number of TUNEL labeled nuclei were determined under microscope for each embryo. Ratio of TUNEL-positive cells to total number of cells was defined as TUNEL-stained nuclei. TUNEL staining for each group was performed at least three times.
ROS Level Assay
Reactive oxygen species levels in blastocysts were determined with 2’,7’ –dichlorofluorescein diacetate (DCFDA, ab113851, Abcam). Live blastocysts were washed 2 times with 1xBuffer. After washing, blastocysts were stained with 20 μM DCFDA at 37°C for 45 min in dark. Blastocysts were washed with 1 x Buffer twice. Then blastocysts were placed in a 50 μL drop of 1 x Buffer covered with Ovoil (10029, Vitrolife). Then, ROS levels were analyzed with Zeiss LSM-880 Airyscan® system (Zeiss, Germany).
Statistical Analysis
All experiment results were analyzed by one-way analysis of variance (one-way ANOVA) followed by Dunn’s post hoc test. We conducted statistical calculations by using SigmaStat for Windows, version 3.5 (Jandel Scientific Corp). For all tests, P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgements
We would like to thank all authors who provided published data.
Abbreviations
Dnmt, DNA methyltransferase ; ICM, inner cell mass ; TE, trophectoderm ; ESC, embryonic stem cell ; ROS, reactive oxygen species ; IF, immunofluorescence ;Declarations
Ethics Approval
The experimental protocol was approved by the Animal Care and Usage Committee of Ankara University (Protocol no: 2022-12-101)
Funding
This study was financially supported by The Scientific and Technological Research Council of Turkey (TUBITAK) 220S736.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author contributions
All authors qualify for authorship by contributing substantially to this article. F.U. and O.C. developed the original concept of this study and wrote the manuscript. All authors collected data and F.U. performed statistical analysis. All authors have contributed to critical discussion, reviewed the final version of the article, and approved it for publication.
Data availability statement
Data are available on request from the corresponding author.
References
Adjaye J., Huntriss J., Herwig R., BenKahla A., Brink T. C., Wierling C., Hultschig C., Groth D., Yaspo M.L., Picton H. M., Gosden R. G., Lehrach H. (2005). Primary Differentiation in the Human Blastocyst: Comparative Molecular Portraits of Inner Cell Mass and Trophectoderm Cells. Stem Cells 23: 1514-1525.
Agarwal A., Aponte-Mellado A., Premkumar B. J., Shaman A., Gupta S. (2012). The effects of oxidative stress on female reproduction: a review. Reproductive Biology and Endocrinology 10: 49.
Assou S., Boumela I., Haouzi D., Monzo C., Dechaud H., Kadoch I.J., Hamamah S. (2012). Transcriptome Analysis during Human Trophectoderm Specification Suggests New Roles of Metabolic and Epigenetic Genes. PLoS ONE 7: e39306.
Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Research 21: 381-395.
Cao J., Yan Q. (2012). Histone Ubiquitination and Deubiquitination in Transcription, DNA Damage Response, and Cancer. Frontiers in Oncology 2: 26.
Cui M., Wen Z., Chen J., Yang Z., Zhang H. (2010). 5-Aza-2′-deoxycytidine is a potent inhibitor of DNA methyltransferase 3B and induces apoptosis in human endometrial cancer cell lines with the up-regulation of hMLH1. Medical Oncology 27: 278-285.
Du J., Johnson L. M., Jacobsen S. E., Patel D. J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nature Reviews Molecular Cell Biology 16: 519-532.
Fatemi M., Hermann A., Gowher H., Jeltsch A. (2002). Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA . European Journal of Biochemistry 269: 4981-4984.
Galán A., Montaner D., Póo M. E., Valbuena D., Ruiz V., Aguilar C., Dopazo J., Simón C. (2010). Functional Genomics of 5- to 8-Cell Stage Human Embryos by Blastomere Single-Cell cDNA Analysis. PLoS ONE 5: e13615.
Gao S., Chung Y. G., Williams J. W., Riley J., Moley K., Latham K. E. (2003). Somatic Cell-Like Features of Cloned Mouse Embryos Prepared with Cultured Myoblast Nuclei1. Biology of Reproduction 69: 48-56.
Gawriluk T. R., Ko C.M., Hong X., Christenson L. K., Rucker E. B. (2014). Beclin-1 deficiency in the murine ovary results in the reduction of progesterone production to promote preterm labor. Proceedings of the National Academy of Sciences 111: E4194-E4203.
Grubelnik G., Boštjančič E., Pavlič A., Kos M., Zidar N. (2020). NANOG expression in human development and cancerogenesis. Experimental Biology and Medicine 245: 456-464.
Habibi R., Hosseini S. M., Zadegan F. G., Hajian M., Ostadhosseini S., Vash N. T., Naddafpour A., Nasr Esfahani M. H. (2018). Functional characterization of NANOG in goat pre-implantation embryonic development. Theriogenology 120: 33-39.
Harvey A.J., Armant D.R., Bavister B.D., Nichols S.M., Brenner C.A. (2009). Inner Cell Mass Localization of NANOG Precedes OCT3/4 in Rhesus Monkey Blastocysts. Stem Cells and Development 18: 1451-1458.
He H., Wang J., Mou X., Liu X., Li Q., Zhong M., Luo B., Yu Z., Zhang J., Xu T., Dou C., Wu D., Qing W., Wu L., Zhou K., Fan Z., Wang T., Hu T., Zhang X., Zhou J., Miao Y.L. (2023). Selective autophagic degradation of ACLY (ATP citrate lyase) maintains citrate homeostasis and promotes oocyte maturation. Autophagy 19: 163-179.
Huang P., Zhou Y., Tang W., Ren C., Jiang A., Wang X., Qian X., Zhou Z., Gong A. (2022). Long-term treatment of Nicotinamide mononucleotide improved age-related diminished ovary reserve through enhancing the mitophagy level of granulosa cells in mice. The Journal of Nutritional Biochemistry 101: 108911.
Kim S., Eun H., Jo E.K. (2019). Roles of Autophagy-Related Genes in the Pathogenesis of Inflammatory Bowel Disease. Cells 8: 77.
Kornicka K., Marycz K., Marędziak M., Tomaszewski K. A., Nicpoń J. (2017). The effects of the DNA methyltranfserases inhibitor 5‐Azacitidine on ageing, oxidative stress and DNA methylation of adipose derived stem cells . Journal of Cellular and Molecular Medicine 21: 387-401.
Lee M., Nam H. Y., Kang H.B., Lee W. H., Lee G.H., Sung G.J., Han M. W., Cho K.J., Chang E.J., Choi K.C., Kim S. W., Kim S. Y. (2021). Epigenetic regulation of p62/SQSTM1 overcomes the radioresistance of head and neck cancer cells via autophagy-dependent senescence induction. Cell Death & Disease 12: 250.
Li X., Zhou L., Peng G., Liao M., Zhang L., Hu H., Long L., Tang X., Qu H., Shao J., Zheng H., Long M. (2021). Pituitary P62 deficiency leads to female infertility by impairing luteinizing hormone production. Experimental & Molecular Medicine 53: 1238-1249.
Liu W. J., Ye L., Huang W. F., Guo L. J., Xu Z. G., Wu H. L., Yang C., Liu H. F. (2016). p62 links the autophagy pathway and the ubiqutin–proteasome system upon ubiquitinated protein degradation. Cellular & Molecular Biology Letters 21: 29.
Luo Z., Xu X., Sho T., Zhang J., Xu W., Yao J., Xu J. (2019). ROS-induced autophagy regulates porcine trophectoderm cell apoptosis, proliferation, and differentiation. American Journal of Physiology-Cell Physiology 316: C198-C209.
Ng R. K., Dean W., Dawson C., Lucifero D., Madeja Z., Reik W., Hemberger M. (2008). Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nature Cell Biology 10: 1280-1290.
Niemann H., Wrenzycki C. (2000). Alterations of expression of developmentally important genes in preimplantation bovine embryos by in vitro culture conditions: Implications for subsequent development. Theriogenology 53: 21-34.
Petrussa L., Van de Velde H., De Rycke M. (2014). Dynamic regulation of DNA methyltransferases in human oocytes and preimplantation embryos after assisted reproductive technologies. MHR: Basic science of reproductive medicine 20: 861-874.
Rossant J. (2004). Lineage development and polar asymmetries in the peri-implantation mouse blastocyst. Seminars in Cell & Developmental Biology 15: 573-581.
Roth Z. (2018). Symposium review: Reduction in oocyte developmental competence by stress is associated with alterations in mitochondrial function. Journal of Dairy Science 101: 3642-3654.
Saitou M., Kagiwada S., Kurimoto K. (2012). Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development 139: 15-31.
Schultz R. M., Williams C. J. (2002). The Science of ART. Science 296: 2188-2190.
Smith Z. D., Meissner A. (2013). DNA methylation: roles in mammalian development. Nature Reviews Genetics 14: 204-220.
Song Z.H., Yu H.Y., Wang P., Mao G.K., Liu W.X., Li M.N., Wang H.N., Shang Y.L., Liu C., Xu Z.L., Sun Q.Y., Li W. (2015). Germ cell-specific Atg7 knockout results in primary ovarian insufficiency in female mice. Cell Death & Disease 6: e1589-e1589.
Sozen B., Can A., Demir N. (2014). Cell fate regulation during preimplantation development: A view of adhesion-linked molecular interactions. Developmental Biology 395: 73-83.
Sritanaudomchai H., Sparman M., Tachibana M., Clepper L., Woodward J., Gokhale S., Wolf D., Hennebold J., Hurlbut W., Grompe M., Mitalipov S. (2009). CDX2 in the formation of the trophectoderm lineage in primate embryos. Developmental Biology 335: 179-187.
Turek-Plewa J., Jagodzinski P. P., (2005). The role of mammalian DNA methyltransferases in the regulation of gene expression. Cellular and Molecular Biology Letters 10: 631-647.
Uysal F., Akkoyunlu G., Ozturk S. (2015). Dynamic expression of DNA methyltransferases (DNMTs) in oocytes and early embryos. Biochimie 116: 103-113.
Uysal F., Cinar O., Can A. (2021). Knockdown of Dnmt1 and Dnmt3a gene expression disrupts preimplantation embryo development through global DNA methylation. Journal of Assisted Reproduction and Genetics 38: 3135-3144.
Uysal F., Ozturk S., Akkoyunlu G. (2017). DNMT1, DNMT3A and DNMT3B proteins are differently expressed in mouse oocytes and early embryos. Journal of Molecular Histology 48: 417-426.
Vivaldi A., Miasaki F.Y., Ciampi R., Agate L., Collecchi P., Capodanno A., Pinchera A., Elisei R. (2009). Re-differentiation of thyroid carcinoma cell lines treated with 5-Aza-2′-deoxycytidine and retinoic acid. Molecular and Cellular Endocrinology 307: 142-148.
Wang L., Tang J., Wang L., Tan F., Song H., Zhou J., Li F. (2021). Oxidative stress in oocyte aging and female reproduction. Journal of Cellular Physiology 236: 7966-7983.
Watson A. J. (2001). Regulation of blastocyst formation. Frontiers in Bioscience 6: d708-730.
Xiong H., Chen Z.F., Liang Q.C., Du W., Chen H.M., Su W.Y., Chen G.Q., Han Z.G., Fang J.Y. (2009). Inhibition of DNA methyltransferase induces G2 cell cycle arrest and apoptosis in human colorectal cancer cells via inhibition of JAK2/STAT3/STAT5 signalling. Journal of Cellular and Molecular Medicine 13: 3668-3679.
XU Y.N., CUI X.S., SUN S.C., LEE S.E., LI Y.H., KWON J.S., LEE S.H., HWANG K.C., KIM N.H. (2011). Mitochondrial Dysfunction Influences Apoptosis and Autophagy in Porcine Parthenotes Developing In Vitro. Journal of Reproduction and Development 57: 143-150.
Xue Z., Huang K., Cai C., Cai L., Jiang C., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y. E., Liu J., Horvath S., Fan G. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500: 593-597.
Yamanaka Y., Ralston A., Stephenson R. O., Rossant J. (2006). Cell and molecular regulation of the mouse blastocyst. Developmental Dynamics 235: 2301-2314.
Zhang J., Tam W.L., Tong G. Q., Wu Q., Chan H.Y., Soh B.S., Lou Y., Yang J., Ma Y., Chai L., Ng H.H., Lufkin T., Robson P., Lim B. (2006). Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nature Cell Biology 8: 1114-1123.
Zhang W., Li K., Zhong X., Yan H., Tong G. (2018). CDX2 is essential for human IVF early embryonic development. Acta Biochimica et Biophysica Sinica 50: 1274-1279.
Zhang Y., Yang Z., Wu J. (2007). Signaling pathways and preimplantation development of mammalian embryos. FEBS Journal 274: 4349-4359.
Zhao D.C., Li Y.M., Ma J.L., Yi N., Yao Z.Y., Li Y.P., Quan Y., Li X.N., Xu C.L., Qiu Y., Wu L.Q. (2019). Single-cell RNA sequencing reveals distinct gene expression patterns in glucose metabolism of human preimplantation embryos. Reproduction, Fertility and Development 31: 237.
Zhou J., He H., Zhang J.J., Liu X., Yao W., Li C., Xu T., Yin S.Y., Wu D.Y., Dou C.L., Li Q., Xiang J., Xiong W.J., Wang L.Y., Tang J.M., Xue Z., Zhang X., Miao Y.L. (2022). ATG7-mediated autophagy facilitates embryonic stem cell exit from naive pluripotency and marks commitment to differentiation. Autophagy 18: 2946-2968.