Int. J. Dev. Biol. 68: 103 - 116 (2024)
The Genetic Odyssey of Axolotl Regeneration: Insights and Innovations
Open Access | Review | Published: 12 December 2024
Abstract
The axolotl, a legendary creature with the potential to regenerate complex body parts, is positioned as a powerful model organism due to its extraordinary regenerative capabilities. Axolotl can undergo successful regeneration of multiple structures, providing us with the opportunity to understand the factors that exhibit altered activity between regenerative and non-regenerative animals. This comprehensive review will explore the mysteries of axolotl regeneration, from the initial cellular triggers to the intricate signaling cascades that guide this complex process. We will delve deeply into the multifaceted interplay of genes and factors, highlighting the key role of signaling pathways and the influence of epigenetic modifications (such as DNA methylation, histone modification, and miRNA regulation) during regeneration. Furthermore, we will discuss how axolotls defy the odds by showing remarkable resistance to cancer, offering insights into potential therapeutic strategies. However, that is not the end; we will also highlight how age might affect the regenerative power of this creature. We hope this review will help navigate the awe-inspiring realm of axolotl regeneration, advance our understanding of regenerative biology, and chart pathways for future investigations aimed at uncovering new therapeutic approaches.
Keywords
Regeneration, Axolotl, Epigenetic modifications, Aging, Cancer
Introduction
Regeneration can be described as the restoration of lost cells, tissues, or organs in organisms and it is thought to be an adaptive capability that organisms have developed in the course of evolution (El Agha et al., 2016). It is considered an evolutionary trait that is distributed unevenly among organs and organisms across the animal kingdom (Somorjai et al., 2012). As we move from the lower level (Invertebrates) to the higher level (vertebrates), there is a substantial decrease in regeneration abilities. Invertebrates such as planarians (Rink, 2013) and Hydra (Vogg et al., 2019) show extensive regeneration abilities and can regenerate entire organisms from small tissue fragments, whereas primitive vertebrates show modest regeneration (Zhao et al., 2016). Among vertebrates, urodeles amphibians like newts and salamanders occupy first rank as vertebrate models to study regeneration, followed by frogs and fish that also have noteworthy regenerative capacities (Gesslbauer and Radtke, 2018). Ambystoma mexicanum (axolotl) is a urodeles amphibian native of a clear water lake in Mexico City named Lake Xochimilco (Humphrey, 1975). Axolotl is considered to be the champion of regeneration as axolotl has mastered the ability to repair or replace tissues after injury or amputation (Roy and Gatien, 2008). This amazing model helps to investigate mechanisms controlling regeneration and cellular behavior to give desired outcomes and pattern formation during the regeneration of limbs (Zhulyn et al., 2023)), gills (Saito et al., 2019), tail (Carbonell M. et al., 2022), lens and also internal structures like heart (Pedersen et al., 2021), brain (Yin et al., 2022) and lungs (Jensen et al., 2021). It can undergo complete and faithful regeneration of complex structures and gives us hope to enhance the regenerative potential in humans (Huang et al., 2024).
Axolotl emerged as an excellent model due to its elegant and unique methods of regenerating lost or injured tissues or organs. It is also considered an excellent model to use in research due to its less evolutionary distance to mammals in the evolutionary tree of life compared to invertebrate models of regeneration. Axolotl belongs to the class Amphibia, which diverged 400 million years ago (Biscotti et al., 2020), and mammals diverged nearly 129 million years ago, while invertebrates like amphioxus diverged 625 million years ago (Igawa et al., 2017). Previously, the molecular techniques used for studying axolotl were limited to biochemical, embryonic and phenomenological studies, and the absence of fully sequenced genome was a major limitation. However, after complete genome sequencing of axolotl, many advanced molecular tools have been developed to identify highly conserved genes and pathways that might be involved in the successful restoration of lost body parts (Sámano et al., 2021; Smith et al., 2005). The RNA sequencing technologies and microarray studies that were being used previously for transcriptional studies can be coupled with the now available genomic sequence of axolotl to gain a complete understanding of gene expression during regeneration.
Furthermore, RNA sequencing at single-cell resolution (Leigh et al., 2018) and cellular reprogramming (Atsuta et al., 2024) will help us to understand cellular diversity, cellular dynamics, and cell lineage and mark the donor tissues. Genomic manipulation of axolotl using CRISPR–Cas9 to create genetically modified axolotl will help to study the function of specific genes in the regeneration process by inserting or removing (knock in or knock out) specific genes (Fei et al., 2018; Tilley et al., 2022). Similarly, gene overexpression or gene knockdown by Electroporation-based method is also being used to study the regeneration process in axolotl by coupling it with gene editing or gene regulatory techniques (Fu et al., 2023). Genetic and epigenetic factors precisely regulate the regeneration process and play an essential role in key regenerative events, so epigenetic control involving DNA methylation, histone modifications, and microRNAs is also emerging as a key area of this research field (Hayashi et al., 2020; Voss et al., 2021; Gupta et al., 2023).
Axolotl as a model: charting regenerative research milestones
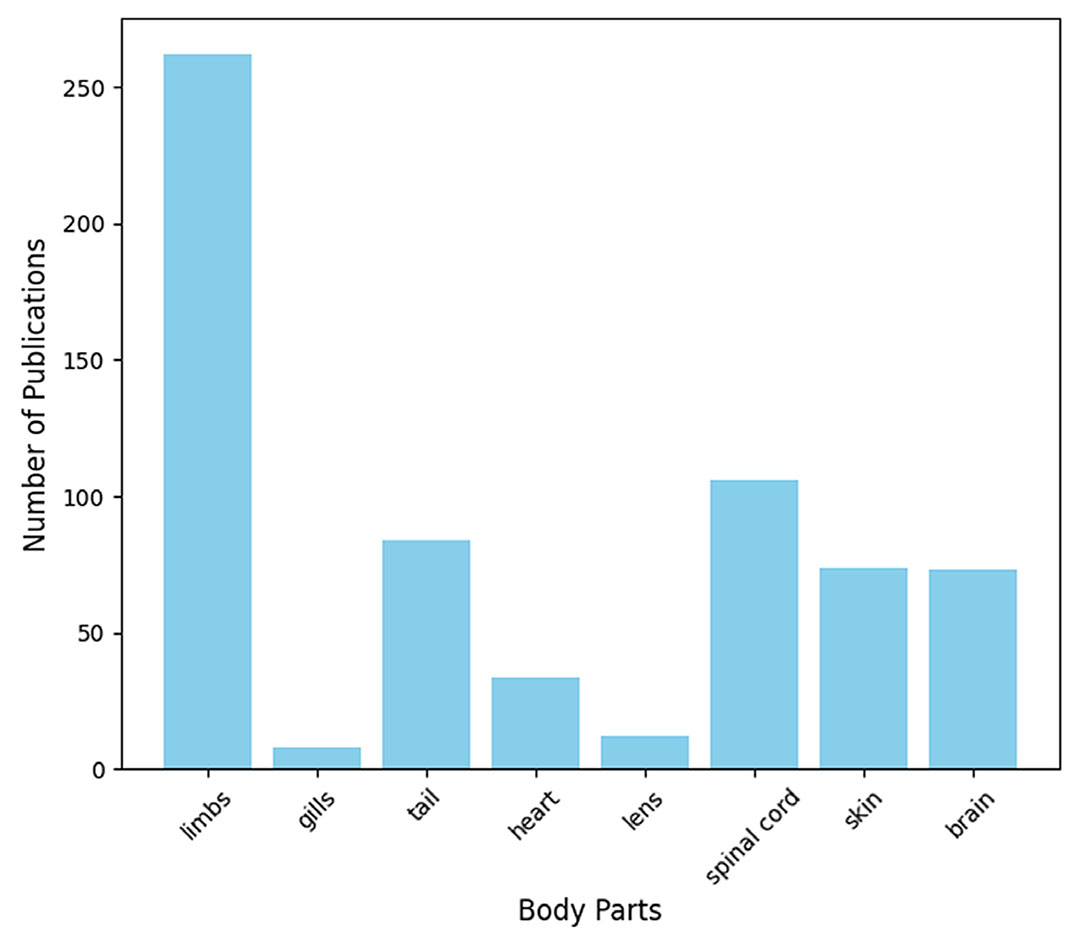
Compared to mammals, in which regenerative capacities are limited to a few parts of the body such as skeletal muscle in adult mammals (Sastourné-Arrey et al., 2023), heart regeneration in neonatal rats (Wang et al., 2020), digits tips of adult mouse (Johnson et al., 2020), and spiny mice that can regenerate cartilage, muscles, dermis, hair follicles, and improved cardiac repair (Peng et al., 2021; Tomasso et al., 2024), axolotl can undergo highly successful regeneration of various structures. Axolotl employs epimorphic mode of regeneration, characterized by the appearance of proliferative blastema at the site of injury or wound and leads to the restoration of the complete structure by subsequent differentiation (Laplace-Builhé et al., 2021). Axolotl has a long history of being used as a research tool to understand and investigate the complex mechanisms of regeneration. Axolotl came into the limelight in 1768 with the discovery of tail and limb regeneration by Spallanzani (1769). Later on, it was cultivated in the laboratory since 1864 to investigate various mechanisms (Reiß, 2022). Until now, the inability to carry out genetic studies and incompletely defined cellular mechanisms in model species possessing the capability to regenerate were the main limitations to answering the fundamental questions of Regenerative Biology. However, recent advancements in molecular technologies have made it possible to overcome these limitations by investigating the regenerative capabilities of model organisms. Axolotl genome size ranges from 14 to 120 GB. A great achievement has been made by sequencing and assembling a large-sized genome of A. mexicanum containing 32 GB distributed in 14 chromosome pairs. It was challenging due to the largest size ever sequenced (10-fold as large as the size of the human genome), where a significant majority, comprising 70%, of the genome consists of repetitive elements (Nowoshilow et al., 2018). Later on, it was validated by mapping it with the transcriptomic data of 22 tissues from different body parts of an axolotl, which ended in 85% alignment. Furthermore, transcriptomic data provided extensive gene datasets and expression profiles specific to tissues. Due to its striking regenerative potential, it has become an ideal organism for research and has gained the attention of many researchers who want to study the hidden mechanisms of regeneration. To get an estimate of the research on axolotl’s regenerative power, we undertook a study on PubMed. We searched with keywords “regeneration” and “axolotl” and each body part (limbs, gills, tail, heart, lens, spinal cord, skin, brain) separately between the years 2000 and 2024 and compared the number of publications for each organ in Fig. 2. Studies conducted on limb regeneration of axolotl surpass the other body parts because limb regeneration is a unique potential observed only in axolotl among tetrapods.
Fig. 2. Total number of publications related to regeneration in various body parts of axolotl.
Between the years 2000 and 2024 was searched on PubMed by using keywords “axolotl” and “limbs”, “gills”, “tail”, “heart”, “lens”, “spinal cord”, “skin” or “brain”. The number of publications in which “regeneration” and “axolotl” were mentioned together is 435 compared to the zebrafish regeneration model which was alluded to in 2,946 publications. While total number of publications mentioning “axolotl” was 754, and publications mentioning “zebrafish” were 48,737. Thus, whereas the % of zebrafish papers dealing with regeneration was 6% (2,946/48,737), that of axolotl papers dealing with regeneration was 58% (435/754).
Dive into the cellular and molecular dynamics of Axolotl regeneration
Although various animal models have been widely studied to understand the molecular basis of regeneration, axolotl emerged as an excellent model due to its elegant and unique methods of regenerating lost or injured tissues or organs. Unlike several other amphibious salamanders, axolotl does not necessarily undergo metamorphoses into terrestrial form, maintaining aquatic larval form even after sexual maturity, and due to this trait, they are named neotenic animals (Demircan et al., 2016). Early researchers also named them “slave of the water” in the sense that they do not metamorphose to adapt themselves into terrestrial existence in contrast to the closely related organisms of the genus Ambystoma. One of their closest relatives is Ambystoma tigrinum, which, unlike A. mexicanum, does not retain juvenile traits and metamorphose into terrestrial form and, along the way, lose its fringed gills and caudal fin (Ryan et al., 2009).
Researchers gave different hypotheses to explain why these species can regenerate while others do not. One hypothesis suggests that as axolotls are neotenic and do not complete metamorphosis, retaining juvenile characteristics or embryonic-like characteristics, so they are capable of regenerating their body parts (Tompkins, 1978; Galliot and Ghila, 2010). This hypothesis was supported by the example of the African clawed frog, which shows robust regeneration at larval stages and loses this ability after the initiation of metamorphosis (Suzuki et al., 2006). However, this hypothesis was not accepted because studies on newts which show endogenous metamorphosis (Iten and Bryant, 1973) and axolotl, in which metamorphosis can be induced by activating thyroid hormone signaling, do not correlate with this hypothesis, and it shows that metamorphosis can restrict regeneration in frogs but not in urodeles (Tompkins and Townsend, 1977; Rosenkilde et al., 1982). Another hypothesis states that the depleted or simple immune system of Urodeles, in contrast to that of mammals, makes them able to regenerate their body parts (Mescher and Neff, 2005). Increased regenerative abilities in some tetrapod species with simpler immune responses support this hypothesis (Godwin and Brockes, 2006). While, other conflicting examples also exist, with developed immunity playing a role in regeneration (Godwin and Brockes, 2006).
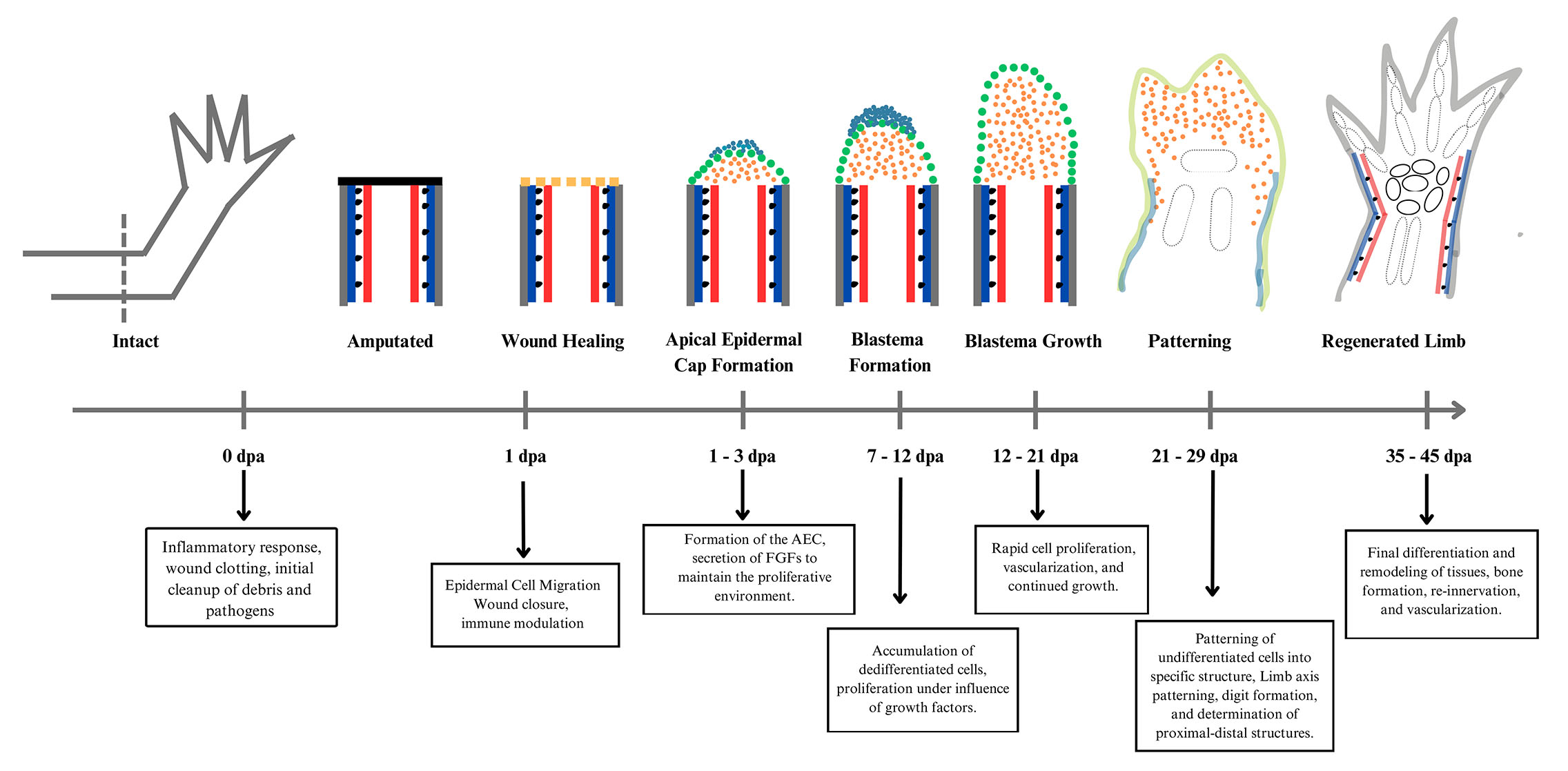
Although axolotls are unique in their ability to regenerate several body parts, limb regeneration in axolotls has been extensively investigated to understand the cellular and molecular mechanisms that regulate tissue or organ regeneration. Limb regeneration in axolotls from amputation to the complete restoration of the lost structure in completed in few distinct stages as described in Fig. 3..
Amputation or injury leading to wound healing
When an animal gets injured or amputated, the underlying tissues become exposed to pathogens, and the injury site becomes full of debris or dead cells. So, several biological processes must be activated to protect from pathogens, mitigate blood loss and clean the debris (Godwin et al., 2013). Immune cells are key players in the wound healing process after injury. Transcriptomic studies have shown that the number of macrophages and neutrophils increases at the amputation site, where neutrophils activate anti-inflammatory macrophages by suppressing NF-κB signalling and macrophages halt the inflammatory response (Rodgers et al., 2020; Marwick et al., 2018; Li et al., 2021). The study of Godwin et al., (2017) indicated that the presence of macrophages is crucial to regulating fibroblast activity and preventing excess fibrosis during the early stages of heart regeneration in axolotl. They also showed that the absence of macrophages at the start of the wound healing process results in excessive fibrosis, altered enzyme activity in extracellular matrix, and impaired regeneration. Neutrophils aim to clean the cellular and molecular debris at the wound site and direct the production of matrix metalloproteinase for collagen degradation. Following amputation or injury, exposed mesenchymal tissues are covered by migrating epithelial cells. Closure of wound is rapid, and it is usually closed in four hours in young axolotl (this timing may increase with age) (Carlson, 1998). This wound healing without scar formation prevents damage, infection, or inflammation (Harty et al., 2003). In mammals, wound healing results in scar formation, which discriminates the wound healing process between mammals and axolotls. It is known that, except for major nerves, their absence does not affect wound healing. At this stage, the wound epidermis is also established, which is critical for triggering the expression of several genes that will participate in events leading to limb regeneration. And if it is removed or disturbed, regeneration will not proceed (Roy and Lévesque, 2006; Goršič, 2007).
Dedifferentiation resulting in blastema formation
In the coming days, the apical epidermal cap, also known as the signaling network, is established to generate various signaling molecules that will help in the dedifferentiation and proliferation of underlying tissues into the limb progenitor cells called blastemal cells (Gardiner et al., 1999; Goršič, 2007). To understand the cellular mechanisms underlying dedifferentiation and blastema formation, Lin et al., (2021) studied the significant differences regarding cellular behaviour and dedifferentiation between the regeneration process of frogs and axolotl. To trace the detailed lineage, they established Prrx1:CreER;CAGGs:lp-Cherry stable transgenic frog lines and observed that blastema cells in frogs do not dedifferentiate into progenitors, resulting in incomplete patterning and regeneration. While in the axolotl, they found complete dedifferentiation and transition to progenitors, highlighting the importance of cellular dedifferentiation. Actually, this stage is characterized by the dedifferentiation of cells in the mature limb tissues, losing their function, migrating under the wound epidermis to accumulate at the tip, and leading to proliferation, finally generating blastema (Bryant et al., 2002). There are several factors that have been reported to regulate blastema formation, for example, Fgf8 and Bmp7(Satoh et al., 2016), Hdac1 (Wang et al., 2019), p53 (Yun et al., 2013), and SHH (Furukawa et al., 2022). The factors affecting blastemal formation are not yet clear, but it has been observed that it depends on the signal generated during the wound healing process. Denervation leads to the failure of blastemal formation, indicating that nerves are required for this stage as it can stop the cells from dedifferentiating (Satoh et al., 2007; Ferretti and Géraudie, 1998).
Redevelopment
During this stage, the undifferentiated mass of cells that were generated during the second stage begins to differentiate, and cells reorganize to restore the lost organ or structure. Cells proliferate, and the blastemal grows and behaves like a developing limb (Muneoka and Bryant, 1982). Nerve dependency decreases in this stage, but blastemal growth still depends on the presence of nerves, and the absence of nerves will result in the failure to regenerate new structures (Stocum, 2011). Apart from that, cellular senescence is induced within the blastema, which helps in regeneration by generating a pro-proliferative environment that stimulates the expansion of neighbouring progenitor cells and supports the overall growth of the blastema (Yu et al., 2023). Although the signals generated from the wound epidermis, apical epidermal cap, or nerves are enough to trigger blastema formation, the growth of blastema requires positional information from the cells that originate from opposite sides of the regenerating limb (Endo et al., 2004). Previous research illustrates that a complete array of positional cues composed of cells from various axes present under the base of the blastema is necessary for regenerating the limb. Molecular profiling and tracking of cells by using transgenic axolotl strains and single-cell RNA sequencing is now being used to understand cell lineage progress during blastema formation and subsequent blastema growth. It helps track individual cell types and the specific gene activity within the growing blastema (Gerber et al., 2018; Currie et al., 2016). Similarly, Super‑paramagnetic iron oxide particles (SPIOs) have also been used to track labelled cells in the regenerating environment of the axolotl limb (Lauridsen et al., 2018). Any missing positional information can be filled by the interaction of cells with each other located in different regions or with different positional information, known as intercalation (Bryant et al., 1981; McCusker et al., 2015).
Influential role of signalling pathways/networks in regeneration
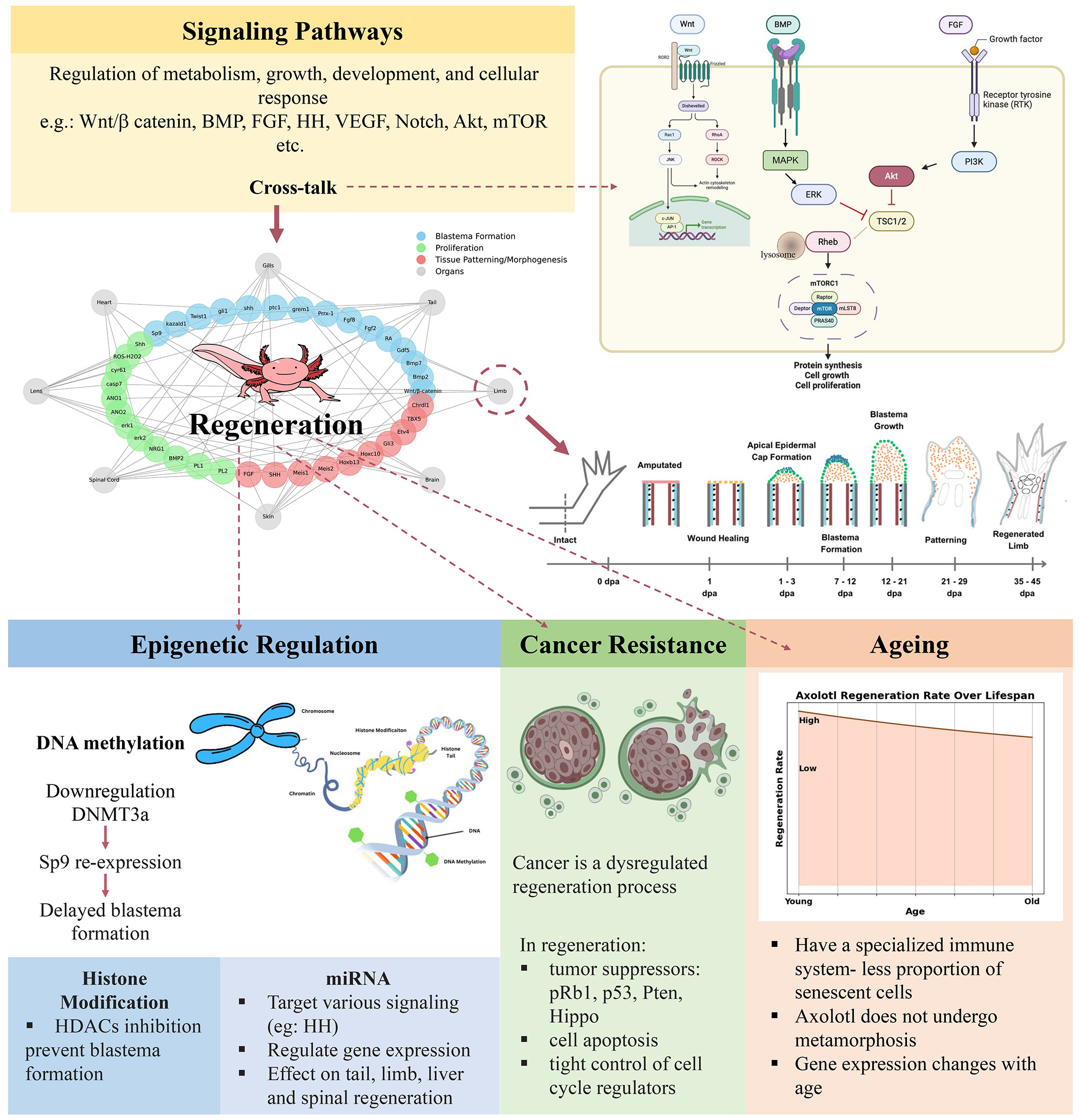
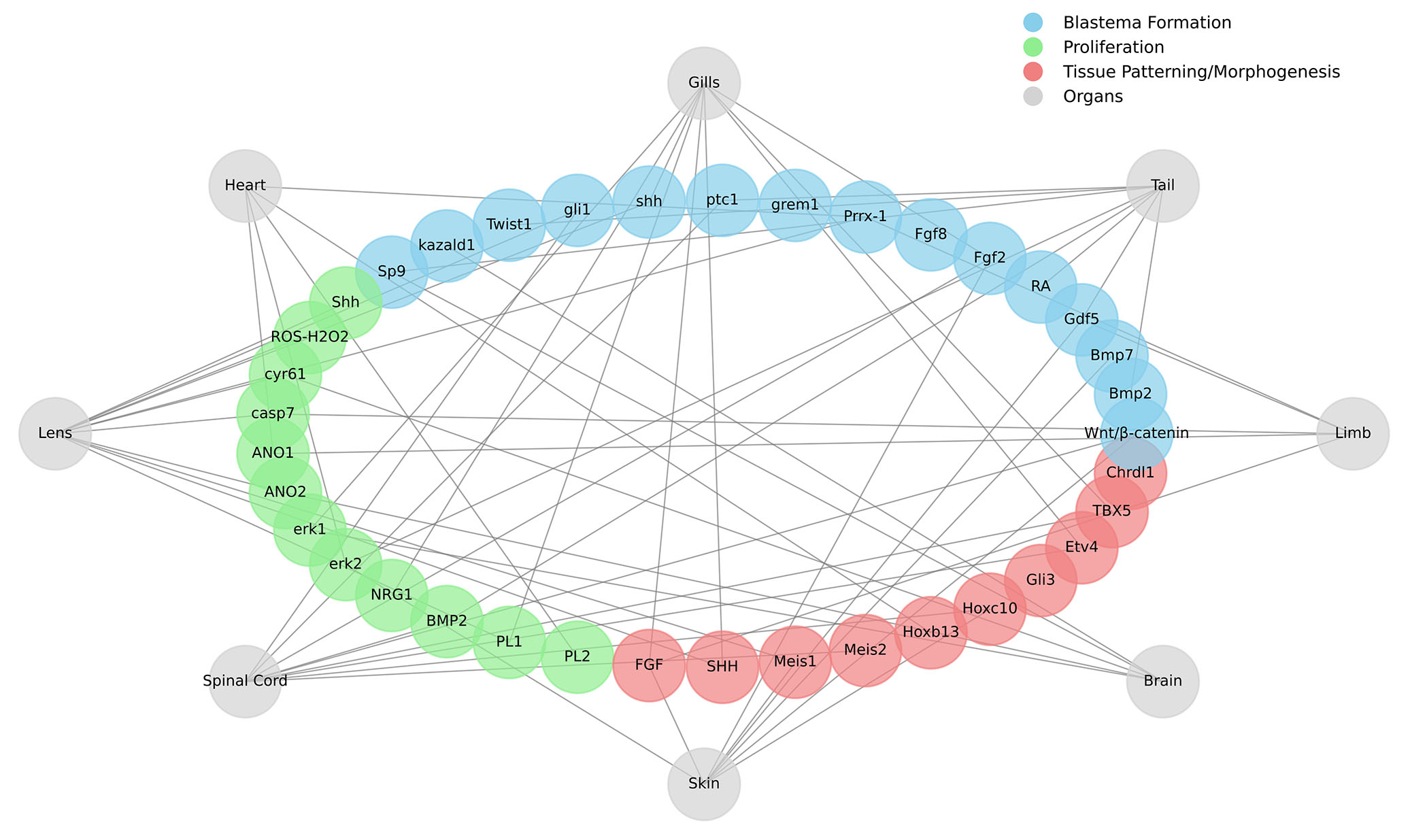
After complete genome sequencing of axolotl, many advanced molecular tools are developed to identify highly conserved genes and pathways that might be involved in the successful restoration of lost body parts (Sámano et al., 2021; Smith et al., 2005). Until now, various signaling pathways and genes have been identified as critical for making regeneration successful, but the function of many genes is still not clearly understood (Sanor et al., 2020). The remarkable ability of axolotl to regenerate its injured or lost body parts is regulated by multiple signaling pathways/factors/genes, which enable the intricate process of growth and repair (Fig. 4). These pathways or signaling networks modulate the cellular responses in a complex way by up-regulation or down-regulation of multiple genes in response to injury or damage. These pathways precisely regulate key regenerative events such as cell differentiation, proliferation, and patterning. The activation of any response inside the cell is carefully organized. Also, these pathways or networks are interconnected to each other, and a signal travelling through one pathway may activate another pathway. So, there is cross-talk or cross-communication between different pathways where activation/termination of one signaling pathway can influence the other pathway until the relevant function is carried out. Many intracellular signaling pathways participating in the regulation of metabolism, growth, development, and cellular response to environmental changes have been studied and the activity of individual proteins involved in these pathways has been investigated (Pryciak, 2009; Kanehisa et al., 2004). For example, the activation of Wnt/β-catenin signaling is crucial for regenerating different body parts of the axolotl, and chemically blocking the Wnt signaling can stop blastema formation (Wischin et al., 2017). Many other genes, factors or pathways have been reported to regulate the blastema formation in axolotl such as Bmp2, Bmp7, Gdf5 (Makanae et al., 2013), RA (Vieira et al., 2019), Fgf2, Fgf8 (Makanae et al., 2014), Prrx-1 (Satoh et al., 2011), grem1, ptc1, shh, gli1, twist1, kazald1 (Bryant et al., 2017), and Sp9 (Satoh et al., 2008). Similarly, the proliferation stage in axolotl regeneration is regulated by several genes such as Shh (Schnapp et al., 2005), ROS especially H2O2 (Carbonell M. et al., 2022), cyr61, casp7 (Voss et al., 2018), Anoctamin1, Anoctamin2, erk1, erk2 (Franklin et al., 2017), NRG1 (Farkas et al., 2016), BMP2 (Lehrberg and Gardiner, 2015), PL1 and PL2 (Zhu et al., 2012). Finally, some genes or factors regulate tissue patterning or morphogenesis such as FGF, SHH (Nacu et al., 2016), RA (Polvadore and Maden, 2021), Meis1, Meis2 (Mercader et al., 2005), Hoxb13, Hoxc10 (Carlson et al., 2001), Gli3, Etv4 (Bickelmann et al., 2018), TBX5, Chrdl1 (Vieira et al., 2023).
Apart from the above described specific stages of regeneration, researchers have found that, the overall regeneration of various body parts of axolotl such as limbs, gill, tail, lens, spinal cord, and skin, is driven by a significant array of genes and signaling pathways, including TGF- β signaling (Lévesque et al., 2007; Ponomareva et al., 2015), Vegf (Ritenour and Dickie, 2017), Notch Signaling (Sousounis et al., 2014; Ponomareva et al., 2015), Akt Signaling (Hayashi et al., 2014), HSP-70 (Lévesque et al., 2005), Yap1 (Bay et al., 2023; Yin et al., 2023; Hayashi et al., 2014), Sox-9 (Guimond et al., 2010), mTOR signaling (Zhulyn et al., 2023), Mmp-9 (Satoh, Hirata and Satou, 2011; Yang et al., 1999), Msx1, Msx2, Gremlin1(Nacu et al., 2016), Wnt-5a, Wnt-5b, and Wnt-7a (Ghosh et al., 2008; Ponomareva et al., 2015), FGF and Bmp (Satoh et al., 2016: Nacu et al., 2016; Satoh et al., 2011; Vieira et al., 2019), HH signaling (Torok et al., 1999; Singh et al., 2018).
Failure in the proper regulation of these mechanisms will lead to the inadequate activation of cellular responses, causing disease (Paulovich et al., 1997). For example, it has been observed that mutation of epidermal growth factor receptor (EGFR) results in various cancer types (Paez et al., 2004). Similarly, malfunction of the Wnt signaling pathway has been investigated to cause colorectal cancer (CRC) (Shimizu et al., 2002). Cell signaling is critical for examining the growth and activity of aberrant cells or cells combatting unfavorable circumstances. To understand the importance of the signaling pathway in any biological process, several experimental strategies are adopted, such as the expression pattern of genes specific to the pathway after using knock-out or knock-down techniques, and resulting effects on the phenotype of cells are observed (Summerton, 2007). Activation or deactivation of genes that code for proteins results in various changes in one or more signaling pathways, and it ultimately alters the cellular response.
Epigenetic regulation of regeneration in Axolotl
Contrary to mammals, axolotl can regenerate its injured or damaged body parts without scar formation and is widely used for understanding the mechanism of regeneration due to its striking regenerative potential. Significant advances have been made in understanding the signaling pathways and cell sources involved in regenerative mechanisms in axolotl. In the field of regenerative biology, epigenetic regulation has gained a central focus, and it involves multiple components such as DNA methylation, chromatin remodelling complexes, miRNAs, and histone modification (Rosa-Garrido et al., 2018). As we know, restoration of any organ is completed through a series of biological responses such as wound healing, cell dedifferentiation, proliferation, and redifferentiation, both genetic and epigenetic alterations trigger transcriptional and translational activities at the molecular level to complete these responses.
Role of DNA methylation in Axolotl regeneration
DNA methylation, which results in the chemical modification of DNA, has a critical role in silencing genes, inactivation of X-chromosomes, genome stability, and the site of methylation in genomic DNA can be the 5th carbon position of 5-cytosine phosphate-guanine-3 dinucleotide (Zhu et al., 2018). Until now, it was believed that DNA methylation results in the inhibition of gene expression, but recent methylation analysis protocols, such as the combination of high-throughput sequencing and whole genome bisulfate sequencing disclosed the other facet of its role in recruiting transcription factors, gene activation, and splicing regulation. In 2015, Aguilar and Gardiner evidenced that the downregulation of DNMT3a expression due to increased nerve signaling plays a role in the early stages of axolotl limb regeneration. They explained that downregulation of DNMT by using beads of decitabine triggers Sp9 re-expression and also results in delayed reformation of the basal lamina, which further aids in blastema formation. If the reformation of basal lamina is not inhibited, it will inhibit the signaling between AEC and blastema mesenchyme leading to failure of blastema formation, so its inhibition is necessary for blastema formation (Neufeld and Day, 1996). Until now, 3 DNA methyltransferases have been spotted, of which DNMT3a and DNMT3b are crucial for de novo methylation of CPG islands while DNMT1 is maintenance methyltransferase (Moore et al., 2013). To maintain the overall status of DNA methylation, methylation and demethylation antagonize each other.
Role of histone modification in Axolotl regeneration
Apart from DNA methylation, chemical changes to histone proteins like acetylation/deacetylation, phosphorylation, or methylation play a crucial role by turning on/off specific genes regulating the regeneration of body parts in axolotl. Actually, these kinds of chemical modifications to histone proteins act like a switchboard that decides which genes are going to be activated or deactivated in any biological response (Zhu et al., 2018). In this type of epigenetic regulation, the histone tail is modified, which results in conformational changes of chromatin, making it loose (euchromatin) or tight (Heterochromatin), leading to gene expression or silencing. Histone acetyltransferases (HATs) and Histone deacetylases (HDACs) have been indicated to accelerate acetylation or deacetylation, respectively. The key role of HDACs is to remodel chromatin to a more condensed form by removing the acetyl group from lysine residues, resulting in gene silencing (Bolden et al., 2006). The study of Wang et al., (2019) showed that using MS-275, which is an HDAC inhibitor prevents blastema formation in the regenerating limb of the axolotl, indicating that HDAC1 expression is necessary for blastema formation. Similar experiments were conducted by Voss et al., (2019) in which they observed the effects of 172 compounds on the tail regeneration of axolotl embryo. Surprisingly, nearly 55 compounds, including histone deacetylase inhibitors (HDACi) such as romidepsin (anti-cancer drug), inhibited tail regeneration in axolotl when treated continuously for seven days, supporting the idea that the activity of HDAC is required for gene expression during regeneration. Previous studies also show that romidepsin treatment results in the overexpression of genes, such as cited2 and txnip, which are negative regulators of Hif1α (Hypoxia-inducible factor-1α), suggesting the association of HDAC activity with oxidative stress and hypoxia (Berlow et al., 2017; Malone et al., 2017). To further validate the inhibitory effects of romidepsin in axolotl tail regeneration, Baddar et al., (2021) used CoCl2, which is a chemical stabilizer of Hif1α (Wagatsuma et al., 2020) together with romidepsin to observe whether CoCl2 can rescue the inhibitory effects romidepsin. They first treated axolotl with romidepsin, which inhibited axolotl tail regeneration and resulted in the high upregulation of cited2. Then, they cotreated axolotl with romidepsin and CoCl2, which partially rescued the inhibitory effects of romidepsin.
Role of miRNA in Axolotl regeneration
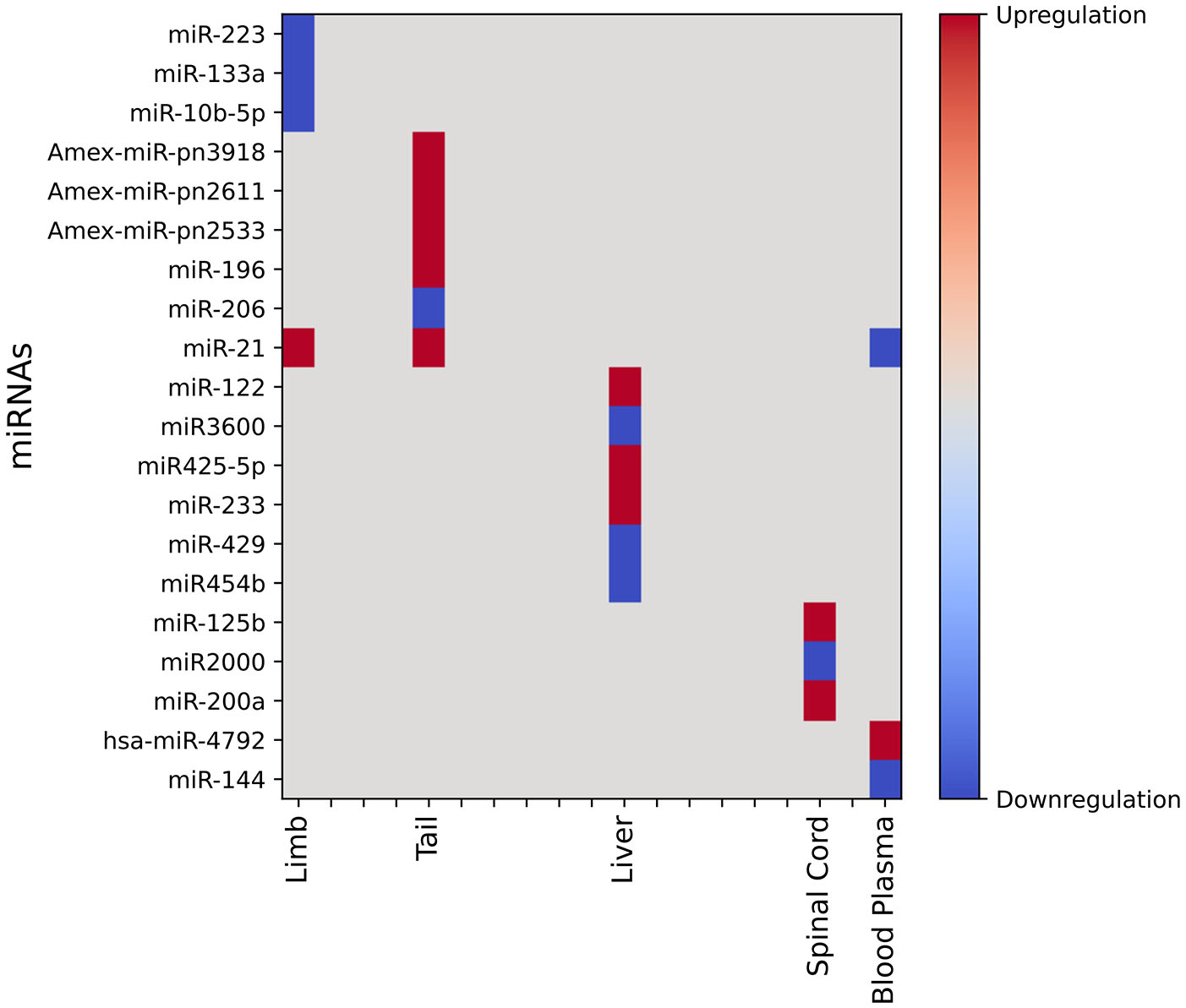
Small non-coding RNAs, also known as miRNAs (microRNAs), degrade the mRNAs by binding to their complementary sequences, resulting in the downregulation of gene expression and, thus, regulating the expression of multiple genes. Many complex biological processes, such as the regeneration of tissues, differentiation, or development, require coordinated gene expression in various cell types. Exploring the key role of miRNAs as effectors of gene expression during regeneration in multiple species, especially Axolotl and Zebrafish, has become an active area of research. These non-coding RNAs are highly conserved, and they can directly or indirectly regulate the regeneration processes by targeting components of various signaling pathways, such as hedgehog or cytokine signaling (Sing et al., 2015). MicroRNAs are versatile molecules that regulate gene expression networks, fine-tune gene expression, cell differentiation and fate, maintain pluripotency of stem cells, cell proliferation, cell reprogramming, and regulate developmental timing (Bartel, 2018). Considering the great regenerative potential of axolotl and the regulatory roles of microRNAs, we provide a summary of recent findings regarding how miRNA regulates regeneration in axolotl in Fig. 5. Sehm et al., (2009) observed the possible role of miR-196 in tail regeneration of axolotl. Direct inhibition of mir-196 leads to defective regeneration and decreased cell proliferation. Another microRNA, miR-206, also plays an essential role in regeneration by suppressing pax7. The suppression of pax7 acts as a signal for the cells to proliferate, and it is necessary for successful regeneration (Gearhart et al., 2015; Sehm et al., 2009). While identifying the novel miRNAs during tail regeneration of axolotl, Gearhart et al., (2015) observed the expression pattern of Amex-miR-pn3918, -pn2611, and –pn2533 in early blastema during tail regeneration and when inhibited defective tail regeneration was witnessed.
A number of miRNAs were also identified to play a critical role during limb regeneration, but this area is poorly understood. Yu et al., (2019) observed the stage-specific regulation of miR-223 and miR-133a. Among them, miR-133a is responsible for regulating the expression of G6PD and SH3G1, while miR-223 specifically regulates the expression of collagen alpha-3 (IX) chain during wound healing or blastema formation. As reported by Holman et al., (2012), the expression of miR-21 was enhanced in the mid-bud blastema during limb regeneration of axolotl, resulting in the downregulation of the Jagged1 gene. Similarly, liver regeneration in axolotl is regulated by multiple miRNAs such as miR-122, miR3600, miR425-5p, and miR-233, which are highly expressed, and miR-429 and miR-454b, which show lower expression in the same organ (Caballero-Pérez et al., 2018). Apart from tail, limb, and liver regeneration in axolotl, the role of miRNAs has also been reported in spinal cord regeneration. Sabin et al., (2019) reported the role of miR-200a in the recovery of spinal cord injury. They found that axolotl glial cells express a non-canonical form of AP-1 complex known as AP-1cFos/JunB, and its overexpression can lead to defects in axon regeneration. To avoid that, glial cells upregulate miR-200a, which inhibits the c-Jun expression and inhibits the formation of AP-1cFos/JunB, resulting in successful regeneration. They also reported that inhibition of miR-200a leads to defective regeneration. Similarly, in another study, the role of miR-200a was identified as the stabilizer of neural stem cell identity during spinal cord regeneration in axolotl. Moreover, its inhibition with an antisense inhibitor changed the fate of cells or caused the neural stem cells to remain in progenitor state (Walker et al., 2022). In another interesting study, Demircan et al., (2021) performed small RNA sequencing on plasma samples of neotenic and metamorphosed axolotls to check the critical role of miRNAs between two groups of axolotls and found 16 differentially expressed circulating miRNAs. They found some miRNAs related to cancer, such as hsa-mir-4792, mir-144, and mir-21, as highly downregulated miRNAs, while p-hsa-mir-304 was highly upregulated. MicroRNAs take part in the regeneration of various body parts in axolotl by regulating the expression pattern of genes and pathways involved in regeneration.
Are Axolotls resistant to cancer?
Apart from astonishing regenerative potential, axolotl shows significant resistance to carcinogens and unlike humans, whose number of cancer cases is increasing every day, axolotl shows an extremely low incidence of cancer (Waddington, 1935). The major risk factors and the mechanism that contributes to the high incidence of cancer are not identified yet but genetic changes and environmental and behavioral factors can be the major contributors. Regeneration and cancer/tumorigenesis share some similar characteristics, but the result of regeneration is life, and cancer results in death as the process of regeneration is strictly controlled by the activity of several tumor suppressors such as pRb1, p53, Pten, and Hippo, while tumorigenesis results in uncontrolled cell proliferation (Charni et al., 2017; Pomerantz and Blau, 2013). Another important factor controlling cell number and proliferation during regeneration to avoid uncontrolled growth is apoptosis, which strictly controls the size of the organ being regenerated (Guerin et al., 2021). Previous studies have also reported that cancer is a dysregulated regeneration process (Schäfer and Werner, 2008) or wounds that do not heal (Dvorak, 2015), even it is also believed that highly regenerative cells have more likelihood of developing cancer (Maggiore and Zhu, 2024).
In axolotls, regeneration permissive environment encompassing both cellular and acellular elements mirrors tumor-like characteristics. For example, the initiation of proliferative signals and the formation of blastema cells, which show phenotypes (proliferation, expression of oncogenes, changes in the chromatin) similar to cancer cells (Vieira et al., 2020). Nevertheless, unlike cancer, the regeneration process in axolotl is well-balanced, and it requires tight control of cell cycle regulators during regeneration (Espinal-Centeno et al., 2020). While in regeneration, the growth of tissues proceeds without fibrosis, but cancer involves fibrosis. Regeneration initiates with the keratinocytes moving towards the wound site to generate an apical epithelium cap (AEC), also known as signaling network after proliferation. Later, it produces various regenerative signaling molecules that are necessary for regeneration (Torres‐Dimas et al., 2022). This regeneration is completely free of any errors or mistakes, which only restores the lost body part. When the regeneration completes, all the signals involved in regulating this complex process must be stopped; otherwise, it will cause abnormal tissue growth and tumor formation (Bölük et al., 2022). Axolotl’s resistance to cancer has also been demonstrated by treatment with various carcinogens like polycyclic hydrocarbons. However, the resistance to carcinogens decreases with the age of axolotl (Ingram, 1971). Suleiman et al., (2020) demonstrated that crude extract from limb tissues of axolotls inhibits cell proliferation, induces differentiation, and modifies gene expression of HL-60 leukemia cells. Additionally, the study of Allegrucci et al., (2011) showed that axolotl oocyte extracts have the potential to induce the reprogramming of breast cancer cells by restoring expressions of silenced tumor suppressors genes, eliminating repressive histone marks on promoters, demethylating DNA and diminishing tumor growth and cancer cell proliferation in mouse xenografts.
Regenerative potential of Axolotl in the face of aging
According to the reports of Illingworth (1974), the rate and efficiency of regeneration in younger people is higher than in old people supporting the idea that age might affect regeneration potential. Humans’ life spans various stages, from embryo to adulthood and finally death, where they go through distinct physiological changes at each stage. Various factors are responsible for these physiological changes, such as nutrition (Shimokawa and Trindade, 2010), cellular metabolism (Afanas’ev, 2010), endocrine signaling (Van Heemst, 2010), genetic or environmental changes (Jin, 2010), etc. Although it is difficult to address the exact stage when aging begins, there are multiple attributes such as adult stem cell pool depletion, a significant decrease in reproductive potential, increased DNA damage and cellular senescence, changes in immune responses, and longer time taken to heal injures, can be linked to aging phenomenon (Jin, 2010). Additionally, increased vulnerability to age-related ailments like heart disease, cancer, and Alzheimer’s disease, decreased potential to self-renew, and capacity to differentiate into progenitor cells are also related to the aging process (Sikora et al., 2010). So, the decrease in regenerative potential in humans due to the aging process is being addressed.
Compared to humans, the life cycle of axolotl is faster, and it takes approximately 9 to 12 months to become sexually mature. There are age-related alterations in the axolotl, such as a decrease in fertility rate (Jones et al., 2014) and changes in the body structure and composition of the skeleton, where the cartilaginous skeleton is replaced by bones with age. In amphibians, ossification starts when they undergo metamorphosis (Quilhac et al., 2014). Riquelme‐Guzmán et al., (2022) showed that, unlike other amphibians, limb skeleton ossification in axolotl continues throughout life with significant transition at the age of 10 months when they reach sexual maturity. They observed a mineralized ring around the cartilaginous mid-diaphysis at the juvenile stage of axolotl while a denser calcified surface around the sexual maturity period. They also observed that ossification can be triggered with exogenous thyroxine. It is reported in other amphibians like Xenopus laevis that regenerative potential decreases significantly after ossification or metamorphosis (Wolfe et al., 2000). Nevertheless, limb regeneration in axolotl persists even after it becomes adult, although it declines with age or after metamorphosis (Monaghan et al., 2014).
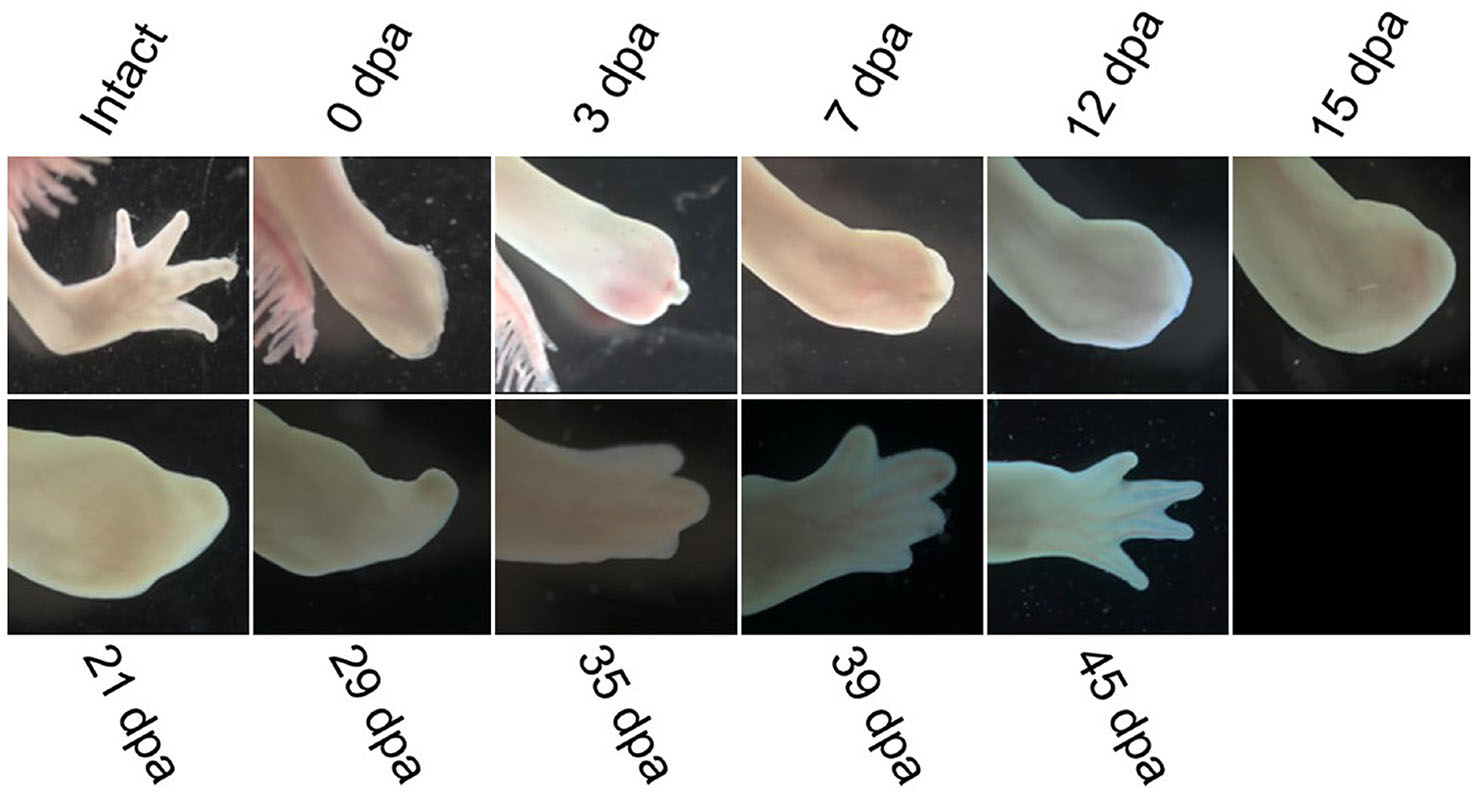
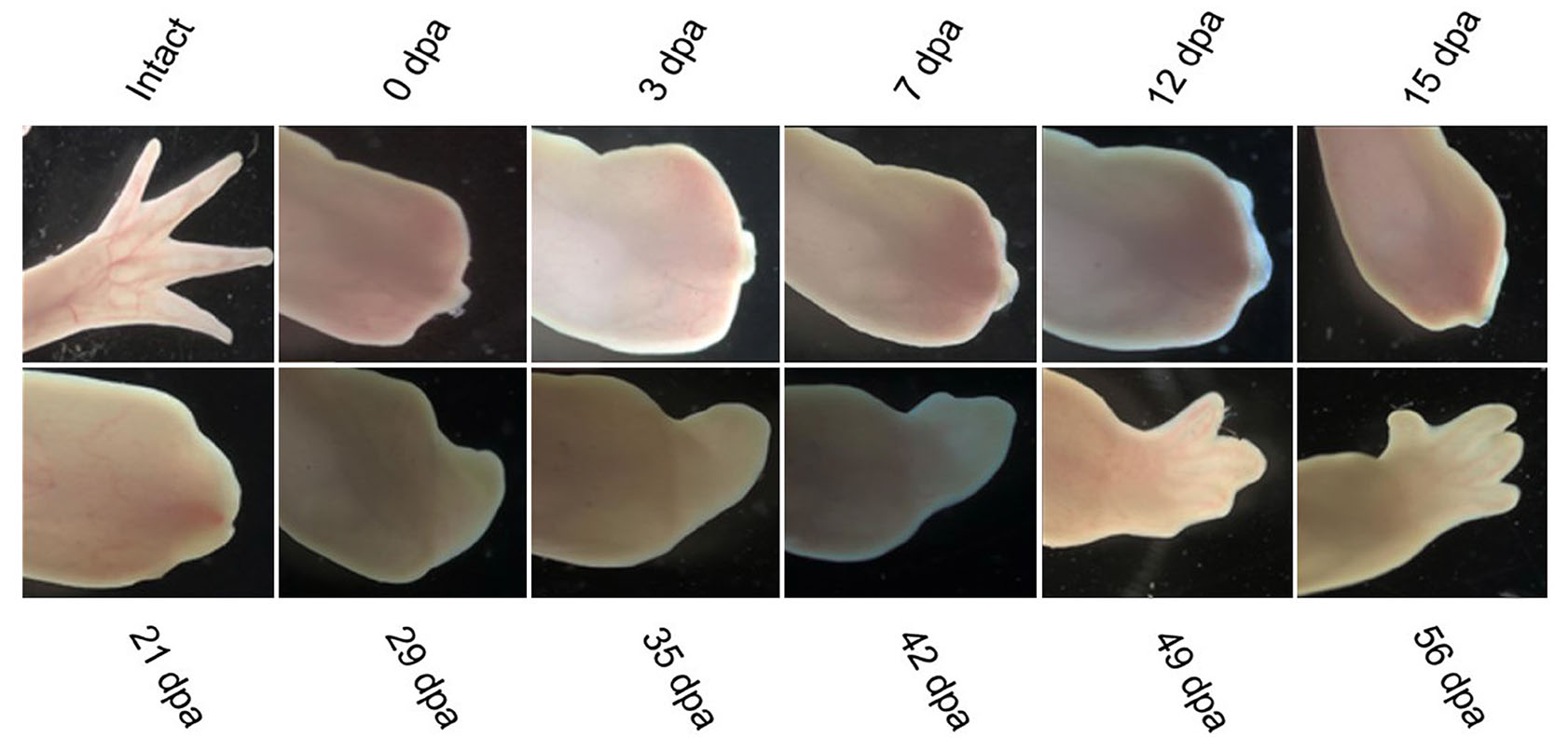
Although it is evident that axolotl ages, these animals show striking resistance to diseases or pathologies that are associated with the aging process and still keep regenerative potential that is way higher than that of humans. Senescent cells that clear damaged or oncogenic cells accumulate in tissues, causing age-related pathologies in humans, and these are also observed in axolotl (Villiard et al., 2017). These cells may be a possible cause of decreased regenerative abilities (Van Deursen, 2014; Herbig et al., 2006). Compared to other animals, axolotls have less proportion of senescent cells in their internal organs as they age, and the level of these cells fluctuates during regeneration, indicating that axolotls exert an immune response to clear senescent cells (Yun et al., 2015). An increase in DNA damage and a decrease in DNA damage response as the animal ages may be another reason behind decreased regenerative potential (Lombard et al., 2005; Sousounis et al., 2020). As the axolotl gets older, its skin becomes thicker, and its flexibility reduces, making it challenging to make wound epithelium in response to injury (Bryant et al., 1981). Furthermore, alterations in the signaling pathways and hormones could cause a decline in regenerative capabilities (Galton, 1992). To get insights into the molecular mechanisms behind regeneration in juvenile and aged axolotls, del Moral-Morales et al., (2023) performed transcriptomic analysis of regenerating limbs and blastema to compare it with old axolotls that could not regenerate successfully. They found a set of differentially expressed genes such as KAZALD1, GPX7, BMP2, WNT5A, WNT5B, DNMT3A, and CTHRC1 in the regenerating tissue of juvenile axolotl while under-expressed in aged axolotl suggesting that these genes have critical role in cell differentiation, cartilage development, bone morphogenesis, transcriptional regulation, and extracellular matrix remodeling. Comparative observations from our laboratory axolotls at two different ages showed that axolotls of 6 months age regenerated faster than those aged 10 months. We selected two axolotls of the same age and the same length of limbs for each group and also amputated at the same length. Fig. 6 shows the regeneration of axolotl at the age of 6 months, and Fig. 7 shows the regeneration of axolotl at the age of 10 months.
Methodology
We selected white mutant (Leucistic) axolotls of age 6 months and 10 months from lab-cultured animals and kept them in separate containers with dechlorinated tap water at 15-20°C. Prior to amputation of limbs, axolotls were anesthetized using 0.1% MS222 solution (ethyl 3-ami-nobenzoate methanesulfonate salt, Sigma-Aldrich, St. Louis, MO). The images were taken for both groups at different regenerative stages by using Zeiss microscope equipped with a color camera Axiocam 208.
Conclusion and future perspective
Among vertebrates, axolotls are endowed with the exceptional ability to restore lost tissues and organs, offering valuable insights into the regeneration mechanism. Although significant progress has been made to find the secrets within its regenerative prowess through the lens of scientific research, our knowledge gained so far is just a piece of a grand mosaic awaiting discoveries. Contrary to the zebrafish model of regeneration, a sparse landscape of publications is a clarion to explore the uncharted territories behind axolotl regeneration. In our review, we have reflected on the wealth of research dedicated to axolotl, emphasizing on various avenues such as the mechanism of regeneration, genetic and epigenetic modifications regulating regeneration, axolotl resistance to cancer, and the impact of age on regenerative potential. The important point that we want to discuss is that the future of axolotl regeneration research is rich with opportunities, giving us the aim to investigate key areas of regeneration by utilizing multidisciplinary approaches such as genomics, epigenomics, proteomics, and transcriptomics, together with advanced technologies. Furthermore, studying axolotl as a regeneration model raises several questions that still need to be answered, such as how feasible it is to transfer the obtained information to the mammalian system or translate the findings of axolotl to species with less regeneration potential as humans. Are there any species-specific factors that help axolotl resist growing tumors upon carcinogen exposure, while humans lack these factors? Are there long-term implications of epigenetic changes on regenerative capacity? If yes, how can we manipulate these changes in other animals to enhance the regenerative potential? Also, axolotl's unique biology or traits limit the generalizability of findings to mammalian species, especially humans. Moreover, as axolotl are endangered in the wild, will their decreasing population pose challenges for ongoing research? The link between regeneration, aging, and cancer warrants deeper research to address the gap in our knowledge and exploit axolotl's transformative potential for regenerative medicines and beyond.
Acknowledgements
The author thanks all the members of his research lab and the readers who gave suggestions for the improvement of the manuscript.
Declarations
Conflicts of interest
The authors declare no conflict of interest.
Funding
This research was funded by the Science & Technology Innovation Project of Laoshan Laboratory (No. LSKJ202203205) and the Open Project of State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University (2023-KF-07).
Ethical approval
This study did not involve any human subjects and was carried out with the approval of Ocean University of China’s Institutional Animal Care and Use Committee.
Data availability
All data supporting the findings of this study are available within the article and it is supplementary material.
References
Afanas'ev I., (2010). Signaling and Damaging Functions of Free Radicals in Aging-Free Radical Theory, Hormesis, and TOR. Aging and Disease 1: 75-88.
Aguilar C., Gardiner D. M., (2015). DNA methylation dynamics regulate the formation of a regenerative wound epithelium during axolotl limb regeneration. PLoS One 10: e0134791.
Allegrucci C., Rushton M. D., Dixon J. E., Sottile V., Shah M., Kumari R., Watson S., Alberio R., Johnson A. D. (2011). Epigenetic reprogramming of breast cancer cells with oocyte extracts. Molecular Cancer 10: 7.
Atsuta Y., Lee C.H., Rodrigues A. R., Colle C., Tomizawa R. R., Lujan E. G., Tschopp P., Galan L., Zhu M., Gorham J. M., Vannier J.P., Seidman C. E., Seidman J. G., Ros M. A., Pourquié O., Tabin C. J. (2024). Direct reprogramming of non-limb fibroblasts to cells with properties of limb progenitors. Developmental Cell 59: 415-430.e8.
Bartel D. P. (2018). Metazoan MicroRNAs. Cell 173: 20-51.
Bay S., Öztürk G., Emekli N., Demircan T. (2023). Downregulation of Yap1 during limb regeneration results in defective bone formation in axolotl. Developmental Biology 500: 31-39.
Berlow R. B., Dyson H. J., Wright P. E. (2017). Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature 543: 447-451.
Bickelmann C., Frota‐Lima G. N., Triepel S. K., Kawaguchi A., Schneider I., Fröbisch N. B. (2018). Noncanonical Hox , Etv4 , and Gli3 gene activities give insight into unique limb patterning in salamanders. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 330: 138-147.
Biscotti M. A., Carducci F., Barucca M., Gerdol M., Pallavicini A., Schartl M., Canapa A., Adolfi M. C. (2020). The transcriptome of the newt Cynops orientalis provides new insights into evolution and function of sexual gene networks in sarcopterygians. Scientific Reports 10: 5445.
Bolden J. E., Peart M. J., Johnstone R. W. (2006). Anticancer activities of histone deacetylase inhibitors. Nature Reviews Drug Discovery 5: 769-784.
Bryant D. M., Johnson K., DiTommaso T., Tickle T., Couger M. B., Payzin-Dogru D., Lee T. J., Leigh N. D., Kuo T.H., Davis F. G., Bateman J., Bryant S., Guzikowski A. R., Tsai S. L., Coyne S., Ye W. W., Freeman R. M., Peshkin L., Tabin C. J., Regev A., Haas B. J., Whited J. L. (2017). A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Reports 18: 762-776.
Bryant S. V., Endo T., Gardiner D. M., (2002). Vertebrate limb regeneration and the origin of limb stem cells. The International Journal of Developmental Biology 46: 887-896.
Bryant S. V., French V., Bryant P. J. (1981). Distal Regeneration and Symmetry. Science 212: 993-1002.
Bölük A., Yavuz M., Demircan T. (2022). Axolotl: A resourceful vertebrate model for regeneration and beyond. Developmental Dynamics 251: 1914-1933.
Caballero-Pérez J., Espinal-Centeno A., Falcon F., García-Ortega L. F., Curiel-Quesada E., Cruz-Hernández A., Bako L., Chen X., Martínez O., Alberto Arteaga-Vázquez M., Herrera-Estrella L., Cruz-Ramírez A. (2018). Transcriptional landscapes of Axolotl (Ambystoma mexicanum). Developmental Biology 433: 227-239.
Carbonell M. B., Zapata Cardona J., Delgado J. P. (2022). Hydrogen peroxide is necessary during tail regeneration in juvenile axolotl. Developmental Dynamics 251: 1054-1076.
Carlson B. M., (1998). Development and regeneration with special emphasis on the amphibian limb. In Cellular and Molecular Basis of Regeneration. (Ed. Ferreti P., ) (Ed. Géraudie J., ) Wiley, Chichester.
Carlson M. R. J., Komine Y., Bryant S. V., Gardiner D. M. (2001). Expression of Hoxb13 and Hoxc10 in Developing and Regenerating Axolotl Limbs and Tails. Developmental Biology 229: 396-406.
Charni M., Aloni-Grinstein R., Molchadsky A., Rotter V. (2017). p53 on the crossroad between regeneration and cancer. Cell Death & Differentiation 24: 8-14.
Currie J. D., Kawaguchi A., Traspas R. M., Schuez M., Chara O., Tanaka E. M. (2016). Live Imaging of Axolotl Digit Regeneration Reveals Spatiotemporal Choreography of Diverse Connective Tissue Progenitor Pools. Developmental Cell 39: 411-423.
- del Moral-Morales Aylin, Sámano Cynthia, Ocampo-Cervantes José Antonio, Topf Maya, Baumbach Jan, González-Barrios Rodrigo, Soto-Reyes Ernesto, (2023). Key Proteins for Regeneration in A. mexicanum: Transcriptomic Insights from Aged and Juvenile Limbs. bioRxiv Preprint: 2023.09.07.556684 10.1101/2023.09.07.556684:
Demircan T., İlhan A. E., Aytürk N., Yıldırım B., Öztürk G., Keskin İ (2016). A histological atlas of the tissues and organs of neotenic and metamorphosed axolotl. Acta Histochemica 118: 746-759.
Demircan T., Sibai M., Avşaroğlu M. E., Altuntaş E., Ovezmyradov G., (2021). The first report on circulating microRNAs at Pre- and Post-metamorphic stages of axolotls. Gene 768: 145258.
Dvorak H. F. (2015). Tumors: Wounds That Do Not Heal—Redux. Cancer Immunology Research 3: 1-11.
El Agha E., Kosanovic D., Schermuly R. T., Bellusci S. (2016). Role of fibroblast growth factors in organ regeneration and repair. Seminars in Cell & Developmental Biology 53: 76-84.
Endo T., Bryant S. V., Gardiner D. M. (2004). A stepwise model system for limb regeneration. Developmental Biology 270: 135-145.
Espinal-Centeno A., Dipp-Álvarez M., Saldaña C., Bako L., Cruz-Ramírez A. (2020). Conservation analysis of core cell cycle regulators and their transcriptional behavior during limb regeneration in Ambystoma mexicanum. Mechanisms of Development 164: 103651.
Farkas J. E., Freitas P. D., Bryant D. M., Whited J. L., Monaghan J. R. (2016). Neuregulin-1 signaling is essential for nerve-dependent axolotl limb regeneration. Development 143: 2724-2731.
Fei J.F., Lou W. P.K., Knapp D., Murawala P., Gerber T., Taniguchi Y., Nowoshilow S., Khattak S., Tanaka E. M. (2018). Application and optimization of CRISPR–Cas9-mediated genome engineering in axolotl (Ambystoma mexicanum). Nature Protocols 13: 2908-2943.
Ferretti P., Geraudie J., (1998). Cellular and Molecular Basis of Regeneration. From Invertebrates to Humans. John Wiley and Sons, Chichester.
Franklin B. M., Voss S. R., Osborn J. L. (2017). Ion channel signaling influences cellular proliferation and phagocyte activity during axolotl tail regeneration. Mechanisms of Development 146: 42-54.
Fu S., Peng C., Zeng Y.Y., Qiu Y., Liu Y., Fei J.F. (2023). Establishing an Efficient Electroporation-Based Method to Manipulate Target Gene Expression in the Axolotl Brain. Cell Transplantation 32: 9636897231200059.
Furukawa S., Yamamoto S., Kashimoto R., Morishita Y., Satoh A., (2022). Variable Shh and Fgf8 positioning in regenerating axolotl limb guarantees consistent limb morphogenesis in different limb sizes. bioRxiv Preprint: 2022.01.04.475010.
Galliot B., Ghila L. (2010). Cell plasticity in homeostasis and regeneration. Molecular Reproduction and Development 77: 837-855.
Galton V. A. (1992). The role of thyroid hormone in amphibian metamorphosis. Trends in Endocrinology & Metabolism 3: 96-100.
Gardiner D.M., Carlson M.R.J., Roy S. (1999). Towards a functional analysis of limb regeneration. Seminars in Cell & Developmental Biology 10: 385-393.
Gearhart M., Erickson J., Walsh A., Echeverri K. (2015). Identification of Conserved and Novel MicroRNAs during Tail Regeneration in the Mexican Axolotl. International Journal of Molecular Sciences 16: 22046-22061.
Gerber T., Murawala P., Knapp D., Masselink W., Schuez M., Hermann S., Gac-Santel M., Nowoshilow S., Kageyama J., Khattak S., Currie J. D., Camp J. G., Tanaka E. M., Treutlein B. (2018). Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration. Science 362: eaaq0681.
Gesslbauer B., Radtke C. (2018). The Regenerative Capability of the Urodele Amphibians and Its Potential for Plastic Surgery. Annals of Plastic Surgery 81: 511-515.
Ghosh S., Roy S., Séguin C., Bryant S. V., Gardiner D. M. (2008). Analysis of the expression and function of Wnt‐5a and Wnt‐5b in developing and regenerating axolotl ( Ambystoma mexicanum ) limbs. Development, Growth & Differentiation 50: 289-297.
Godwin J. W., Brockes J. P. (2006). Regeneration, tissue injury and the immune response. Journal of Anatomy 209: 423-432.
Godwin J. W., Debuque R., Salimova E., Rosenthal N. A. (2017). Heart regeneration in the salamander relies on macrophage-mediated control of fibroblast activation and the extracellular landscape. npj Regenerative Medicine 2: 22.
Godwin J. W., Pinto A. R., Rosenthal N. A. (2013). Macrophages are required for adult salamander limb regeneration. Proceedings of the National Academy of Sciences 110: 9415-9420.
Goršič M., (2007). Axolotl—a supermodel for tissue regeneration. Slovenian Veterinary Research 44: 5-9.
Guerin D. J., Kha C. X., Tseng K. A.S. (2021). From Cell Death to Regeneration: Rebuilding After Injury. Frontiers in Cell and Developmental Biology 9: 655048.
Guimond J.C., Lévesque M., Michaud P.L., Berdugo J., Finnson K., Philip A., Roy S. (2010). BMP-2 functions independently of SHH signaling and triggers cell condensation and apoptosis in regenerating axolotl limbs. BMC Developmental Biology 10: 15.
Gupta S., Dutta S., Hui S. P. (2023). Regenerative Potential of Injured Spinal Cord in the Light of Epigenetic Regulation and Modulation. Cells 12: 1694.
Harty M., Neff A. W., King M. W., Mescher A. L. (2003). Regeneration or scarring: An immunologic perspective. Developmental Dynamics 226: 268-279.
Hayashi S., Tamura K., Yokoyama H. (2014). Yap1, transcription regulator in the Hippo signaling pathway, is required for Xenopus limb bud regeneration. Developmental Biology 388: 57-67.
Hayashi S., Tamura K., Yokoyama H. (2020). Chromatin dynamics underlying the precise regeneration of a vertebrate limb – Epigenetic regulation and cellular memory. Seminars in Cell & Developmental Biology 97: 16-25.
Herbig U., Ferreira M., Condel L., Carey D., Sedivy J. M. (2006). Cellular Senescence in Aging Primates. Science 311: 1257-1257.
Holman E. C., Campbell L. J., Hines J., Crews C. M. (2012). Microarray Analysis of microRNA Expression during Axolotl Limb Regeneration. PLoS ONE 7: e41804.
Huang L., Ho C., Ye X., Gao Y., Guo W., Chen J., Sun J., Wen D., Liu Y., Liu Y., Zhang Y., Li Q. (2024). Mechanisms and translational applications of regeneration in limbs: From renewable animals to humans. Annals of Anatomy - Anatomischer Anzeiger 255: 152288.
Humphrey R. R., (1975). The axolotl, Ambystoma mexicanum. In Handbook of Genetics. Springer, Boston.
Igawa T., Nozawa M., Suzuki D. G., Reimer J. D., Morov A. R., Wang Y., Henmi Y., Yasui K. (2017). Evolutionary history of the extant amphioxus lineage with shallow-branching diversification. Scientific Reports 7: 1157.
Illingworth C. M., (1974). Trapped fingers and amputated finger tips in children. Journal of Pediatric Surgery 9: 853-858.
Ingram A. J. (1971). The reactions to carcinogens in the axolotl ( Ambystoma mexicanum ) in relation to the ‘regeneration field control’ hypothesis. Development 26: 425-441.
Iten L. E., Bryant S. V., (1973). Forelimb regeneration from different levels of amputation in the newt, Notophthalmus viridescens: Length, rate, and stages. Wilhelm Roux' Archiv für Entwicklungsmechanik der Organismen 173: 263-282.
Jensen T. B., Giunta P., Schultz N. G., Griffiths J. M., Duerr T. J., Kyeremateng Y., Wong H., Adesina A., Monaghan J. R. (2021). Lung injury in axolotl salamanders induces an organ‐wide proliferation response. Developmental Dynamics 250: 866-879.
Jin K., (2010). Modern Biological Theories of Aging. Aging and Disease 1: 72-74.
Johnson G. L., Masias E. J., Lehoczky J. A. (2020). Cellular Heterogeneity and Lineage Restriction during Mouse Digit Tip Regeneration at Single-Cell Resolution. Developmental Cell 52: 525-540.e5.
Jones O. R., Scheuerlein A., Salguero-Gómez R., Camarda C. G., Schaible R., Casper B. B., Dahlgren J. P., Ehrlén J., García M. B., Menges E. S., Quintana-Ascencio P. F., Caswell H., Baudisch A., Vaupel J. W. (2014). Diversity of ageing across the tree of life. Nature 505: 169-173.
Kanehisa M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Research 32: 277D-280.
Kawakami Y., Rodriguez Esteban C., Raya M., Kawakami H., Martí M., Dubova I., Izpisúa Belmonte J. C. (2006). Wnt/β-catenin signaling regulates vertebrate limb regeneration. Genes & Development 20: 3232-3237.
Kim W.S., Stocum D. L. (1986). Effects of retinoids on regenerating limbs: comparison of retinoic acid and arotinoid at different amputation levels. Roux's Archives of Developmental Biology 195: 455-463.
Laplace-Builhé B., Bahraoui S., Jorgensen C., Djouad F. (2021). From the Basis of Epimorphic Regeneration to Enhanced Regenerative Therapies. Frontiers in Cell and Developmental Biology 8: 605120.
Lauridsen H., Foldager C., Hansen L., Pedersen M. (2018). Non-invasive cell tracking of SPIO labeled cells in an intrinsic regenerative environment: The axolotl limb. Experimental and Therapeutic Medicine 15: 3311-3319.
Lehrberg J., Gardiner D. M. (2015). Regulation of Axolotl (Ambystoma mexicanum) Limb Blastema Cell Proliferation by Nerves and BMP2 in Organotypic Slice Culture. PLOS ONE 10: e0123186.
Leigh N. D., Dunlap G. S., Johnson K., Mariano R., Oshiro R., Wong A. Y., Bryant D. M., Miller B. M., Ratner A., Chen A., Ye W. W., Haas B. J., Whited J. L. (2018). Transcriptomic landscape of the blastema niche in regenerating adult axolotl limbs at single-cell resolution. Nature Communications 9: 5153.
Li H., Wei X., Zhou L., Zhang W., Wang C., Guo Y., Li D., Chen J., Liu T., Zhang Y., Ma S., Wang C., Tan F., Xu J., Liu Y., Yuan Y., Chen L., Wang Q., Qu J., Shen Y., Liu S., Fan G., Liu L., Liu X., Hou Y., Liu G.H., Gu Y., Xu X. (2021). Dynamic cell transition and immune response landscapes of axolotl limb regeneration revealed by single-cell analysis. Protein & Cell 12: 57-66.
Lin T.Y., Gerber T., Taniguchi-Sugiura Y., Murawala P., Hermann S., Grosser L., Shibata E., Treutlein B., Tanaka E. M. (2021). Fibroblast dedifferentiation as a determinant of successful regeneration. Developmental Cell 56: 1541-1551.e6.
Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., Alt F. W. (2005). DNA Repair, Genome Stability, and Aging. Cell 120: 497-512.
Lévesque M., Gatien S., Finnson K., Desmeules S., Villiard E., Pilote M., Philip A., Roy S. (2007). Transforming Growth Factor: β Signaling Is Essential for Limb Regeneration in Axolotls. PLoS ONE 2: e1227.
Lévesque M., Guimond J.C., Pilote M., Leclerc S., Moldovan F., Roy S. (2005). Expression of heat‐shock protein 70 during limb development and regeneration in the axolotl. Developmental Dynamics 233: 1525-1534.
Maggiore G., Zhu H. (2024). Relationships Between Regeneration, Wound Healing, and Cancer. Annual Review of Cancer Biology 8: 177-197.
Makanae A., Hirata A., Honjo Y., Mitogawa K., Satoh A. (2013). Nerve independent limb induction in axolotls. Developmental Biology 381: 213-226.
Makanae A., Mitogawa K., Satoh A. (2014). Co-operative Bmp- and Fgf-signaling inputs convert skin wound healing to limb formation in urodele amphibians. Developmental Biology 396: 57-66.
Malone C. F., Emerson C., Ingraham R., Barbosa W., Guerra S., Yoon H., Liu L. L., Michor F., Haigis M., Macleod K. F., Maertens O., Cichowski K. (2017). mTOR and HDAC Inhibitors Converge on the TXNIP/Thioredoxin Pathway to Cause Catastrophic Oxidative Stress and Regression of RAS-Driven Tumors. Cancer Discovery 7: 1450-1463.
Marwick J. A., Mills R., Kay O., Michail K., Stephen J., Rossi A. G., Dransfield I., Hirani N. (2018). Neutrophils induce macrophage anti-inflammatory reprogramming by suppressing NF-κB activation. Cell Death & Disease 9: 665.
McCusker C., Bryant S. V., Gardiner D. M. (2015). The axolotl limb blastema: cellular and molecular mechanisms driving blastema formation and limb regeneration in tetrapods. Regeneration 2: 54-71.
Mercader N., Tanaka E. M., Torres M. (2005). Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development 132: 4131-4142.
Mescher A. L., Neff A. W. (2005). Regenerative Capacity and the Developing Immune System. In Regenerative Medicine I. (Ed. Yannas I. V.) Springer Berlin Heidelberg, Berlin, Heidelberg.
Monaghan J. R., Stier A. C., Michonneau F., Smith M. D., Pasch B., Maden M., Seifert A. W. (2014). Experimentally induced metamorphosis in axolotls reduces regenerative rate and fidelity. Regeneration 1: 2-14.
Moore L. D., Le T., Fan G. (2013). DNA Methylation and Its Basic Function. Neuropsychopharmacology 38: 23-38.
Muneoka K., Bryant S. V. (1982). Evidence that patterning mechanisms in developing and regenerating limbs are the same. Nature 298: 369-371.
Nacu E., Gromberg E., Oliveira C. R., Drechsel D., Tanaka E. M. (2016). FGF8 and SHH substitute for anterior–posterior tissue interactions to induce limb regeneration. Nature 533: 407-410.
Neufeld D. A., Day F. A. (1996). Perspective: A suggested role for basement membrane structures during newt limb regeneration. The Anatomical Record 246: 155-161.
Nowoshilow S., Schloissnig S., Fei J.F., Dahl A., Pang A. W. C., Pippel M., Winkler S., Hastie A. R., Young G., Roscito J. G., Falcon F., Knapp D., Powell S., Cruz A., Cao H., Habermann B., Hiller M., Tanaka E. M., Myers E. W. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature 554: 50-55.
Paez J. G., Jänne P. A., Lee J. C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F. J., Lindeman N., Boggon T. J., Naoki K., Sasaki H., Fujii Y., Eck M. J., Sellers W. R., Johnson B. E., Meyerson M. (2004). EGFR Mutations in Lung Cancer: Correlation with Clinical Response to Gefitinib Therapy. Science 304: 1497-1500.
Paulovich A. G., Toczyski D. P., Hartwell L. H. (1997). When Checkpoints Fail. Cell 88: 315-321.
Pedersen K., Rasmussen R. K., Dittrich A., Lauridsen H. (2021). Cardiac regeneration in the axolotl is unaffected by alterations in leukocyte numbers induced by lipopolysaccharide and prednisolone. BMC Research Notes 14: 157.
Peng H., Shindo K., Donahue R. R., Gao E., Ahern B. M., Levitan B. M., Tripathi H., Powell D., Noor A., Elmore G. A., Satin J., Seifert A. W., Abdel-Latif A. (2021). Adult spiny mice (Acomys) exhibit endogenous cardiac recovery in response to myocardial infarction. npj Regenerative Medicine 6: 74.
Polvadore T., Maden M. (2021). Retinoic Acid Receptors and the Control of Positional Information in the Regenerating Axolotl Limb. Cells 10: 2174.
Pomerantz J. H., Blau H. M. (2013). Tumor suppressors: enhancers or suppressors of regeneration?. Development 140: 2502-2512.
Ponomareva L. V., Athippozhy A., Thorson J. S., Voss S. R. (2015). Using Ambystoma mexicanum (Mexican axolotl) embryos, chemical genetics, and microarray analysis to identify signaling pathways associated with tissue regeneration. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology 178: 128-135.
Pryciak P. M. (2009). Designing New Cellular Signaling Pathways. Chemistry & Biology 16: 249-254.
Quilhac A., de Ricqlès A., Lamrous H., Zylberberg L. (2014). Globuli ossei in the long limb bones of Pleurodeles waltl ( Amphibia , Urodela , Salamandridae ). Journal of Morphology 275: 1226-1237.
Reiß C. (2022). Cut and Paste: The Mexican Axolotl, Experimental Practices and the Long History of Regeneration Research in Amphibians, 1864-Present. Frontiers in Cell and Developmental Biology 10: 786533.
Rink J. C. (2013). Stem cell systems and regeneration in planaria. Development Genes and Evolution 223: 67-84.
Riquelme‐Guzmán C., Schuez M., Böhm A., Knapp D., Edwards‐Jorquera S., Ceccarelli A. S., Chara O., Rauner M., Sandoval‐Guzmán T. (2022). Postembryonic development and aging of the appendicular skeleton in Ambystoma mexicanum. Developmental Dynamics 251: 1015-1034.
Ritenour A. M., Dickie R. (2017). Inhibition of Vascular Endothelial Growth Factor Receptor Decreases Regenerative Angiogenesis in Axolotls. The Anatomical Record 300: 2273-2280.
Rodgers A.K., Smith J.J., Voss S.R. (2020). Identification of immune and non-immune cells in regenerating axolotl limbs by single-cell sequencing. Experimental Cell Research 394: 112149.
Rosa-Garrido M., Chapski D. J., Vondriska T. M. (2018). Epigenomes in Cardiovascular Disease. Circulation Research 122: 1586-1607.
Rosenkilde P., Mogensen E., Centervall G., Jørgensen O. S. (1982). Peaks of neuronal membrane antigen and thyroxine in larval development of the Mexican axolotl. General and Comparative Endocrinology 48: 504-514.
Roy S., Gatien S. (2008). Regeneration in axolotls: a model to aim for!. Experimental Gerontology 43: 968-973.
Roy S., Lévesque M. (2006). Limb Regeneration in Axolotl: Is It Superhealing?. The Scientific World JOURNAL 6: 12-25.
Ryan M. E., Johnson J. R., Fitzpatrick B. M. (2009). Invasive hybrid tiger salamander genotypes impact native amphibians. Proceedings of the National Academy of Sciences 106: 11166-11171.
Sabin K. Z., Jiang P., Gearhart M. D., Stewart R., Echeverri K. (2019). AP-1cFos/JunB/miR-200a regulate the pro-regenerative glial cell response during axolotl spinal cord regeneration. Communications Biology 2: 91.
Saito N., Nishimura K., Makanae A., Satoh A. (2019). Fgf- and Bmp-signaling regulate gill regeneration in Ambystoma mexicanum. Developmental Biology 452: 104-113.
Sámano C., González-Barrios R., Castro-Azpíroz M., Torres-García D., Ocampo-Cervantes J. A., Otero-Negrete J., Soto-Reyes E. (2021). Genomics and epigenomics of axolotl regeneration. The International Journal of Developmental Biology 65: 465-474.
Sanor L. D., Flowers G. P., Crews C. M. (2020). Multiplex CRISPR/Cas screen in regenerating haploid limbs of chimeric Axolotls. eLife 9: e48511.
Sastourné-Arrey Q., Mathieu M., Contreras X., Monferran S., Bourlier V., Gil-Ortega M., Murphy E., Laurens C., Varin A., Guissard C., Barreau C., André M., Juin N., Marquès M., Chaput B., Moro C., O’Gorman D., Casteilla L., Girousse A., Sengenès C. (2023). Adipose tissue is a source of regenerative cells that augment the repair of skeletal muscle after injury. Nature Communications 14: 80.
Satoh A., Gardiner D. M., Bryant S. V., Endo T. (2007). Nerve-induced ectopic limb blastemas in the axolotl are equivalent to amputation-induced blastemas. Developmental Biology 312: 231-244.
Satoh A., Graham G.M.C., Bryant S.V., Gardiner D.M. (2008). Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum). Developmental Biology 319: 321-335.
Satoh A., Makanae A., Nishimoto Y., Mitogawa K. (2016). FGF and BMP derived from dorsal root ganglia regulate blastema induction in limb regeneration in Ambystoma mexicanum. Developmental Biology 417: 114-125.
Satoh A., makanae A., Hirata A., Satou Y. (2011). Blastema induction in aneurogenic state and Prrx-1 regulation by MMPs and FGFs in Ambystoma mexicanum limb regeneration. Developmental Biology 355: 263-274.
Schnapp E., Kragl M., Rubin L., Tanaka E. M. (2005). Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development 132: 3243-3253.
Schäfer M., Werner S. (2008). Cancer as an overhealing wound: an old hypothesis revisited. Nature Reviews Molecular Cell Biology 9: 628-638.
Sehm T., Sachse C., Frenzel C., Echeverri K. (2009). miR-196 is an essential early-stage regulator of tail regeneration, upstream of key spinal cord patterning events. Developmental Biology 334: 468-480.
Shimizu Y., Ikeda S., Fujimori M., Kodama S., Nakahara M., Okajima M., Asahara T. (2002). Frequent alterations in the Wnt signaling pathway in colorectal cancer with microsatellite instability. Genes, Chromosomes and Cancer 33: 73-81.
Shimokawa I., Trindade L. S., (2010). Dietary restriction and aging in rodents: a current view on its molecular mechanisms. Aging and Disease 1: 89-107.
Sikora E., Scapagnini G., Barbagallo M. (2010). Curcumin, inflammation, ageing and age-related diseases. Immunity & Ageing 7: 1.
Singh B. N., Koyano-Nakagawa N., Donaldson A., Weaver C., Garry M., Garry D. (2015). Hedgehog Signaling during Appendage Development and Regeneration. Genes 6: 417-435.
Singh B. N., Weaver C. V., Garry M. G., Garry D. J., (2018). Hedgehog and Wnt Signaling Pathways Regulate Tail Regeneration. Stem Cells and Development 27: 1426-1437.
Smith J. J., Putta S., Walker J. A., Kump D. K., Samuels A. K., Monaghan J. R., Weisrock D. W., Staben C., Voss S. R. (2005). Sal-Site: Integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics 6: 181.
Somorjai I. M. L., Somorjai R. L., Garcia-Fernàndez J., Escrivà H. (2012). Vertebrate-like regeneration in the invertebrate chordate amphioxus. Proceedings of the National Academy of Sciences 109: 517-522.
Sousounis K., Athippozhy A. T., Voss S. R., Tsonis P. A. (2014). Plasticity for axolotl lens regeneration is associated with age‐related changes in gene expression. Regeneration 1: 47-57.
Sousounis K., Bryant D. M., Martinez Fernandez J., Eddy S. S., Tsai S. L., Gundberg G. C., Han J., Courtemanche K., Levin M., Whited J. L. (2020). Eya2 promotes cell cycle progression by regulating DNA damage response during vertebrate limb regeneration. eLife 9: e51217.
Spallanzani L., (1769). An essay on animal reproductions. T. Becket and P. A. de Hondt.
Stocum D. L. (2011). The role of peripheral nerves in urodele limb regeneration. European Journal of Neuroscience 34: 908-916.
Suleiman S., Di Fiore R., Cassar A., Formosa M. M., Schembri-Wismayer P., Calleja-Agius J. (2020). Axolotl Ambystoma mexicanum extract induces cell cycle arrest and differentiation in human acute myeloid leukemia HL-60 cells. Tumor Biology 42: 101042832095473.
Summerton J. (2007). Morpholino, siRNA, and S-DNA Compared: Impact of Structure and Mechanism of Action on Off-Target Effects and Sequence Specificity. Current Topics in Medicinal Chemistry 7: 651-660.
Suzuki M., Yakushiji N., Nakada Y., Satoh A., Ide H., Tamura K. (2006). Limb Regeneration in Xenopus laevis Froglet. The Scientific World JOURNAL 6: 26-37.
Tilley L., Papadopoulos S.C., Pende M., Fei J.F., Murawala P. (2022). The use of transgenics in the laboratory axolotl. Developmental Dynamics 251: 942-956.
Tomasso A., Disela V., Longaker M. T., Bartscherer K. (2024). Marvels of spiny mouse regeneration: cellular players and their interactions in restoring tissue architecture in mammals. Current Opinion in Genetics & Development 87: 102228.
Tompkins R. (1978). Genie Control of Axolotl Metamorphosis. American Zoologist 18: 313-319.
Tompkins R., Townsend J. K. (1977). Control of metamorphic events in a neotenous urodele Ambystoma mexicanum. Journal of Experimental Zoology 200: 191-196.
Torok M. A., Gardiner D. M., Izpisúa-Belmonte J. C., Bryant S. V., (1999). Sonic hedgehog (shh) expression in developing and regenerating axolotl limbs. The Journal of Experimental Zoology 284: 197-206.
Torres‐Dimas E., Cruz‐Ramírez A., Bermúdez‐Cruz R. M. (2022). Cancer in Amphibia, a rare phenomenon?. Cell Biology International 46: 1992-1998.
van Deursen J. M. (2014). The role of senescent cells in ageing. Nature 509: 439-446.
van Heemst D., (2010). Insulin, IGF-1 and longevity. Aging and Disease 1: 147-157.
Vieira W. A., Raymond M., Kelley K., Cherubino M. A., Sahin H., McCusker C. D. (2023). Integration failure of regenerated limb tissue is associated with incongruencies in positional information in the Mexican axolotl. Frontiers in Cell and Developmental Biology 11: 1152510.
Vieira W. A., Wells K. M., Raymond M. J., De Souza L., Garcia E., McCusker C. D. (2019). FGF, BMP, and RA signaling are sufficient for the induction of complete limb regeneration from non-regenerating wounds on Ambystoma mexicanum limbs. Developmental Biology 451: 146-157.
Vieira W. A., Wells K. M., McCusker C. D. (2020). Advancements to the Axolotl Model for Regeneration and Aging. Gerontology 66: 212-222.
Villiard É., Denis J.F., Hashemi F. S., Igelmann S., Ferbeyre G., Roy S. (2017). Senescence gives insights into the morphogenetic evolution of anamniotes. Biology Open 891-896.
Vogg M. C., Galliot B., Tsiairis C. D. (2019). Model systems for regeneration: Hydra. Development 146: dev177212.
Randal Voss S., Murrugarra D., Jensen T. B., Monaghan J. R. (2018). Transcriptional correlates of proximal-distal identify and regeneration timing in axolotl limbs. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology 208: 53-63.
Voss S. R., Ponomareva L. V., Dwaraka V. B., Pardue K. E., Baddar N. W. A. H., Rodgers A. K., Woodcock M. R., Qiu Q., Crowner A., Blichmann D., Khatri S., Thorson J. S. (2019). HDAC Regulates Transcription at the Outset of Axolotl Tail Regeneration. Scientific Reports 9: 6751.
Voss S. R., Smith J. J., Cecil R. F., Kabangu M., Duerr T. J., Monaghan J. R., Timoshevskaya N., Ponomareva L. V., Thorson J. S., Veliz-Cuba A., Murrugarra D. (2021). HDAC Inhibitor Titration of Transcription and Axolotl Tail Regeneration. Frontiers in Cell and Developmental Biology 9: 767377.
Waddington C. H. (1935). Cancer and the Theory of Organisers. Nature 135: 606-608.
Wagatsuma A., Arakawa M., Matsumoto H., Matsuda R., Hoshino T., Mabuchi K. (2020). Cobalt chloride, a chemical hypoxia-mimicking agent, suppresses myoblast differentiation by downregulating myogenin expression. Molecular and Cellular Biochemistry 470: 199-214.
Walker S. E., Sabin K. Z., Gearhart M. D., Yamamoto K., Echeverri K. (2022). Regulation of stem cell identity by miR-200a during spinal cord regeneration. Development 149: dev200033.
Wang H., Paulsen M. J., Hironaka C. E., Shin H. S., Farry J. M., Thakore A. D., Jung J., Lucian H. J., Eskandari A., Anilkumar S., Wu M. A., Cabatu M. C., Steele A. N., Stapleton L. M., Zhu Y., Woo Y. J. (2020). Natural Heart Regeneration in a Neonatal Rat Myocardial Infarction Model. Cells 9: 229.
Wang M.H., Wu C.H., Huang T.Y., Sung H.W., Chiou L.L., Lin S.P., Lee H.S. (2019). Nerve-mediated expression of histone deacetylases regulates limb regeneration in axolotls. Developmental Biology 449: 122-131.
Wischin S., Castañeda-Patlán C., Robles-Flores M., Chimal-Monroy J. (2017). Chemical activation of Wnt/β-catenin signalling inhibits innervation and causes skeletal tissue malformations during axolotl limb regeneration. Mechanisms of Development 144: 182-190.
Wolfe A. D., Nye H. L.D., Cameron J. A. (2000). Extent of ossification at the amputation plane is correlated with the decline of blastema formation and regeneration inXenopus laevis hindlimbs. Developmental Dynamics 218: 681-697.
Yang E. V., Gardiner D. M., Carlson M. R.J., Nugas C. A., Bryant S. V. (1999). Expression ofMmp-9 and related matrix metalloproteinase genes during axolotl limb regeneration. Developmental Dynamics 216: 2-9.
Yin B., Li X., Lin G., Wang H. (2022). High-resolution single-cell analysis paves the cellular path for brain regeneration in salamanders. Cell Regeneration 11: 37.
Yin B., Zhang K., Du X., Cai H., Ye T., Wang H. (2023). Developmental switch from morphological replication to compensatory growth for salamander lung regeneration. Cell Proliferation 56: e13369.
Yu Q., Walters H. E., Pasquini G., Pal Singh S., Lachnit M., Oliveira C. R., León-Periñán D., Petzold A., Kesavan P., Subiran Adrados C., Garteizgogeascoa I., Knapp D., Wagner A., Bernardos A., Alfonso M., Nadar G., Graf A. M., Troyanovskiy K. E., Dahl A., Busskamp V., Martínez-Máñez R., Yun M. H. (2023). Cellular senescence promotes progenitor cell expansion during axolotl limb regeneration. Developmental Cell 58: 2416-2427.e7.
Yu Y., Tang J., Su J., Cui J., Xie X., Chen F. (2019). Integrative Analysis of MicroRNAome, Transcriptome, and Proteome during the Limb Regeneration of Cynops orientalis. Journal of Proteome Research 18: 1088-1098.
Yun M. H., Davaapil H., Brockes J. P. (2015). Recurrent turnover of senescent cells during regeneration of a complex structure. eLife 4: e05505.
Yun M. H., Gates P. B., Brockes J. P. (2013). Regulation of p53 is critical for vertebrate limb regeneration. Proceedings of the National Academy of Sciences 110: 17392-17397.
Zhao A., Qin H., Fu X. (2016). What Determines the Regenerative Capacity in Animals?. BioScience 66: 735-746.
Zhu W., Pao G. M., Satoh A., Cummings G., Monaghan J. R., Harkins T. T., Bryant S. V., Randal Voss S., Gardiner D. M., Hunter T. (2012). Activation of germline-specific genes is required for limb regeneration in the Mexican axolotl. Developmental Biology 370: 42-51.
Zhu X., Xiao C., Xiong J.W. (2018). Epigenetic Regulation of Organ Regeneration in Zebrafish. Journal of Cardiovascular Development and Disease 5: 57.
Zhulyn O., Rosenblatt H. D., Shokat L., Dai S., Kuzuoglu-Öztürk D., Zhang Z., Ruggero D., Shokat K. M., Barna M. (2023). Evolutionarily divergent mTOR remodels translatome for tissue regeneration. Nature 620: 163-171.