Int. J. Dev. Biol. 68: 161 - 168 (2024)
Special Issue: Developmental Biology in Nordic Countries
DUX4, the rockstar of embryonic genome activation?
Open Access | Review | Published: 13 May 2024
Abstract
During the initial days of development, the embryo gradually shifts from reliance on maternally provided RNAs and proteins to regulation of its own development. This transition is marked by embryonic genome activation (EGA). While the factors driving human EGA remain poorly characterized, accumulating evidence suggests that double homeobox 4 (DUX4) is an important regulator of this process. Despite advances in single-cell methods which have allowed studies in early human embryos, fundamental questions regarding the function and regulation of DUX4 persist. Here, we review current knowledge of DUX4 with a focus on EGA in humans.
Keywords
Double Homeobox 4 (DUX4), embryo development, embryonic genome activation, gene regulation, non-coding genome
Introduction
The oocyte-to-embryo transition culminates in the degradation of maternal transcripts and in embryonic genome activation (EGA) (Schulz and Harrison, 2019; Vastenhouw et al., 2019). The transcriptome changes prominently during the oocyte-to-four cell and four-to-eight cell transitions that are considered as the minor and major EGA stages, respectively (Braude, 1988; Petropoulos et al., 2016; Tesařék, 1988; Töhönen et al., 2015; Vassena et al., 2011; Xue et al., 2013; Yan et al., 2013). In addition to the activation of key developmental genes, the non-coding genome that contributes to genome regulation becomes extensively transcribed (Bouckenheimer et al., 2016; Paloviita and Vuoristo, 2022). The activation of the embryonic transcriptome is intricately linked with major alterations in the epigenome and chromatin architecture (Chen et al., 2019; Liu et al., 2019; Wu et al., 2018; Xia et al., 2019). It is conceivable that EGA takes place only in a favorable epigenomic landscape that generates adequate conditions for timely gene regulation. How these processes are regulated in human fertilized oocytes and pre-implantation embryos, and whether the factors involved exhibit redundancy, remain poorly understood.
DUX4 belongs to a group of double homeobox genes that are unique to placental mammals. These genes are characterized by two proximal homeoboxes that encode DNA-binding homeodomains (Gabriëls et al., 1999). The primate specific DUX4 is believed to have originated through retro-transposition of the ancestral DUXC gene, followed by the loss of DUXC from the primate genome (Leidenroth et al., 2012). DUX4 mRNA is enriched in human zygotes and cleavage-stage embryos (De Iaco et al., 2017; Hendrickson et al., 2017; Liu et al., 2019; Töhönen et al., 2017; Vuoristo et al., 2022). Silencing of DUX4 in human embryos leads to inefficient degradation of maternal transcripts and incomplete EGA, which implies a potential role of DUX4 as an EGA regulator (Liu et al., 2022; Vuoristo et al., 2022). Human embryonic stem cells (hESCs) have been used to elucidate possible roles of selected EGA factors given the shortage of supernumerary embryos donated for research and the fact that experiments in human embryos are challenging due to ethical and technical limitations (Gawriyski et al., 2023; Hendrickson et al., 2017; Madissoon et al., 2016; Vuoristo et al., 2022; Zou et al., 2022). In hESCs, ectopic expression of DUX4 activates both coding and non-coding genes that are typically active in early human embryo at the time of EGA (Hendrickson et al., 2017; Taubenschmid-Stowers et al., 2022; Vuoristo et al., 2022; Yoshihara et al., 2022). These findings collectively suggest a pivotal role for DUX4 in regulating human EGA. In this review, we aim to provide the latest insight into DUX4 and discuss its significance in the context of human EGA.
The peculiar DUX4 repeat locus
The DUX4 open reading frames are located in the subtelomeric region of chromosome 4, within a macrosatellite repeat region known as D4Z4 (Gabriëls et al., 1999) (Fig. 1). While the D4Z4 repeat array is typically epigenetically repressed in most tissues, it becomes transiently derepressed in human embryos possibly due to global epigenome reprogramming (De Iaco et al., 2017; Hendrickson et al., 2017; Liu et al., 2019; Vuoristo et al., 2022; Xia et al., 2019). The repetitive nature, high GC content, and low expression level of DUX4 pose challenges for sequencing and annotation of this genomic region, particularly when working with human embryos that are available in limited numbers. Most of our current knowledge about DUX4 stems from research on its involvement in the pathogenesis of facioscapulohumeral muscular dystrophy (FSHD) (Campbell et al., 2018). FSHD is caused by derepression of D4Z4 locus which likely leads to a burst of DUX4 expression in a subset of affected muscle cells resulting in cell death (Rickard et al., 2015; Snider et al., 2010). The derepression of D4Z4 locus is caused by either a reduction of D4Z4 repeat units alone (FSHD1), or defects in D4Z4 chromatin repressor SMCHD1 (FSHD2), or both (FSHD1+2) (Hewitt, 2015; Sacconi et al., 2019). Interestingly, a nearly identical D4Z4 repeat array exists on chromosome 10, but the contraction of this repeat array does not cause FSHD presumably due to the lack of a permissive polyadenylation signal (Lemmers et al., 2010). The toxic effect of DUX4 in FSHD pathophysiology is not yet completely understood but the mechanisms likely involve the activation of the MYC-mediated apoptotic pathway and the double-stranded RNA innate immune response pathway (Shadle et al., 2017), as well as repression of nonsense-mediated decay (NMD) (Campbell et al., 2023; Feng et al., 2015; Jagannathan et al., 2019). Notably, DUX4 is cytotoxic not only for muscle cells but also for various other cell types (Kowaljow et al., 2007; Resnick et al., 2019; Rickard et al., 2015; Wallace et al., 2011; Yoshihara et al., 2022). Given that in human embryos MYC expression is only upregulated at the time of major EGA stage, the first two days of development take place without one of the main factors behind DUX4-induced cell death. This could be one of the reasons why human embryos tolerate short-term DUX4 expression.
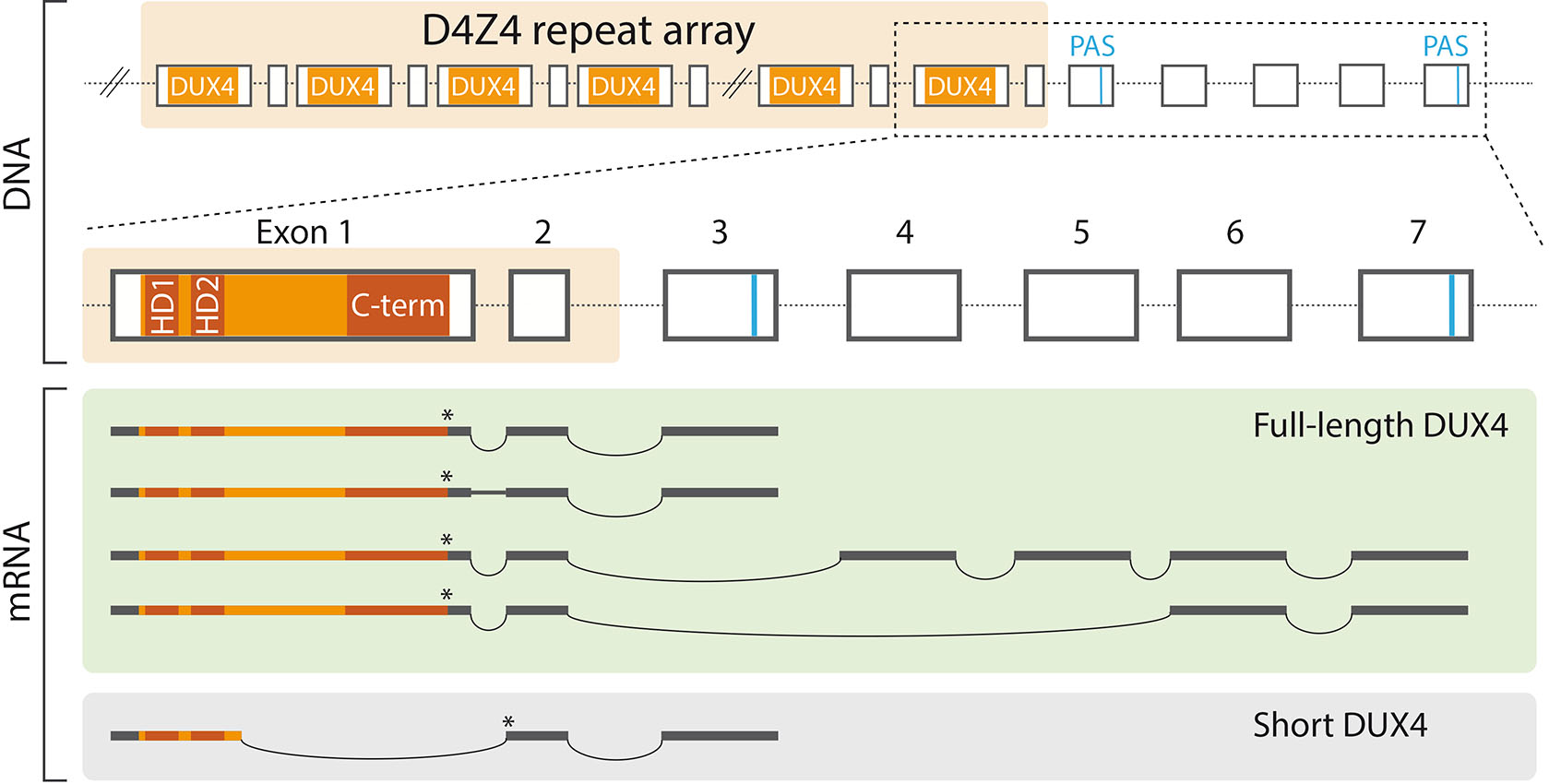
Fig. 1. The D4Z4 repeat array and DUX4 mRNA isoforms.
The protein coding DUX4 transcripts are thought to originate from the last D4Z4 repeat unit and to utilize exons distal to the repeat array that provide canonical polyadenylation signals (PAS). DUX4 transcript isoforms are generated through alternative splicing, but the regulation of this process is poorly understood. In facioscapulohumeral muscular dystrophy (FSHD) muscle, DUX4 mRNA contains exons 1-2-3, and the first intron is alternatively spliced (Snider et al., 2010). Testicular tissue expresses DUX4 mRNA isoforms with exons 1-2-4-5-6-7 and 1-2-6-7 (Snider et al., 2010). Healthy myoblasts use a cryptic splice site within the first exon and express a DUX4 mRNA isoform that is predicted to give rise to a truncated DUX4 (short DUX4 or DUX4-s) that contains the N-terminal homeodomains, but lacks the C-terminus (Snider et al., 2010). Additional DUX4 mRNA isoforms that utilize alternative polyadenylation signals are likely to exist (Smith et al., 2023). The asterisk (*) indicates the location of the translation stop codon. Abbreviations: C-term, C-terminus; DUX4, double homeobox 4; HD, homeodomain; PAS, polyadenylation signal.
Although each repeat unit in the D4Z4 array contains the DUX4 open reading frame, the current conception is that functional DUX4 transcripts originate from the last repeat unit (Fig. 1). This is explained by the position of the polyadenylation signal, which is distal to the repeat array and thus transcribed exclusively as part of the last repeat (Dixit et al., 2007). Consequently, individuals that have a contracted D4Z4 repeat region but lack the distal polyadenylation signal exhibit a normal muscle phenotype (Lemmers et al., 2010). In addition to the DUX4 mRNA isoforms that are transcribed from the D4Z4 array on chromosome 4 and use the conventional polyadenylation signal, numerous isoforms that utilize alternative polyadenylation sites have been described to originate from both chromosome 4 and 10. These isoforms likely result from alternative splicing, however the mechanisms that control the splicing of DUX4 transcripts remain obscure. Moreover, it is unclear which mRNA isoforms give rise to functional DUX4 proteins. Testis and some cancer cell lines express DUX4 transcripts that most likely produce a complete DUX4, as indicated by immunofluorescence stainings and the activation of DUX4 target genes (Smith et al., 2023; Snider et al., 2010). In contrast, various somatic tissues, including healthy muscle, express low levels of capped and polyadenylated DUX4 transcript isoform, which probably produces a truncated protein that lacks the transcription activation domain and, consequently, the ability to activate DUX4 targets (Snider et al., 2010). DUX4 mRNA isoforms present in human embryos have not been described.
The properties of DUX4
The DUX4 protein contains an N-terminal DNA binding domain and C-terminal transcription activation domains (Lee et al., 2021; Mitsuhashi et al., 2018; Vuoristo et al., 2022). The N-terminal DNA-binding domain of DUX4 includes two homeodomains, HD1 and HD2, that arose from an internal duplication of a single homeodomain and consequently, exhibit a high degree of similarity (Leidenroth and Hewitt, 2010). A primate specific mutation changing arginine to glutamate in HD1 have led to the different target DNA sequence preferences of HD1 and HD2; 5′-TAAT-3′ and 5′-TGAT-3′, respectively (Lee et al., 2018). DUX4 has been shown to regulate a set of transposable elements (TEs) (Vuoristo et al., 2022; Young et al., 2013). Hence, the unique HD1 target sequence in primates, as opposed to other mammals, may have evolved as a consequence of co-evolution with species-specific TEs (Lee et al., 2018).
While all the human DUX family members, including DUX4, DUXA and DUXB, contain the N-terminal DNA-binding domain, only DUX4 possesses the conserved C-terminal domain (Leidenroth and Hewitt, 2010). The C-terminus of DUX4 contains a nine amino acid transactivation domain (9aaTAD) (Mitsuhashi et al., 2018) and a KIX-binding motif (KBM) (Vuoristo et al., 2022). The C-terminal domain is essential for DUX4-mediated transcriptional activation and cytotoxicity (Bosnakovski et al., 2008; Choi et al., 2016; Mitsuhashi et al., 2018). Consequently, DUXA and DUXB, which lack the C-terminal domain, are incapable of activating transcription and do not cause cytotoxicity (Bosnakovski et al., 2023). The region between the N-terminal homeodomains and C-terminal transactivating domain is predicted to be intrinsically disordered (Mitsuhashi et al., 2018). While many transcription factors contain an intrinsically disordered region (IDR), the precise nature of their function is not well understood (Ferrie et al., 2022). The deletion of the region between homeodomains and C-terminus does not affect the activation of the known DUX4 target gene ZSCAN4 (Choi et al., 2016; Mitsuhashi et al., 2018) nor does it protect from the DUX4-elicited cell death in myoblasts (Choi et al., 2016) and HEK293 cells (Mitsuhashi et al., 2018). However, further studies are required to determine whether the IDR lacking form of DUX4 can fully recapitulate the function of full-length DUX4.
DUX4 in EGA regulation
DUX4 was first associated with embryogenesis when it was observed to activate early developmental genes in muscle cells of FSHD patients (Geng et al., 2012). Since then, experiments conducted in various cell types, including human myoblasts (Jagannathan et al., 2016; Resnick et al., 2019; Rickard et al., 2015), human pluripotent stem cells (De Iaco et al., 2017; Hendrickson et al., 2017; Taubenschmid-Stowers et al., 2022; Vuoristo et al., 2022; Whiddon et al., 2017; Yoshihara et al., 2022), and most recently various cancer cell lines (Smith et al., 2023), have consistently reported the activation of genes and TEs characteristic of early embryos upon induced or spontaneous DUX4 expression. DUX4 mRNA is enriched in zygotes and early cleavage stage embryos (De Iaco et al., 2017; Liu et al., 2019; Töhönen et al., 2017; Vuoristo et al., 2022). DUX4 protein shows nuclear accumulation in 2- and 4- cell stage embryos followed by rapid clearance by the 8-cell stage (Hendrickson et al., 2017; Vuoristo et al., 2022). Given the ethical and technical challenges associated with research involving human embryos, many insights into the DUX4 functions have been derived from the studies conducted using inducible DUX4 transgene cell lines. Inspired by these findings, the scientific community has recognized DUX4 as one of the earliest regulators of EGA.
DUX4 activates the non-coding genome
In accordance with the timing of the nuclear localization of DUX4 (Vuoristo et al., 2022), the accessible genomic regions in 2-cell and 4-cell embryos are enriched with DUX4 binding motifs (Hendrickson et al., 2017; Liu et al., 2019; Wu et al., 2018). Accessible chromatin regions in human cleavage stage embryos are frequently located at distal sites (> 5 kb from the transcriptional start sites) of genes and enriched at TEs (Gao et al., 2018; Liu et al., 2019; Wu et al., 2018). TEs are mobile genetic elements constituting approximately half of the human genome (Franke et al., 2017). TEs are enormously diverse and divided into distinct classes and families based on their modes of transposition, structural features, and evolutionary relationship (Bourque et al., 2018). TEs promote genetic diversity by moving within the genome, which can lead to alterations in host genes and regulatory sequences. This mobility poses a significant risk to genome stability, and therefore several mechanisms have evolved to suppress TEs (Almeida et al., 2022).
Human TEs are transcriptionally and post-transcriptionally repressed by various chromatin remodellers, Krüppel-associated box (KRAB) domain-containing zinc finger proteins, and small RNAs (Almeida et al., 2022; Gainetdinov et al., 2017; Janssen et al., 2018). However, a substantial proportion of TEs, including human endogenous retroviral (HERV) elements, are transiently active during pre-implantation development (DiRusso and Clark, 2023; Göke et al., 2015; Grow et al., 2015; Liu et al., 2019; Pontis et al., 2019; Xu et al., 2022). For instance, MLT2A1 elements that are members of the HERV family, become accessible and transcribed at the 4-cell stage, retain these states for the 8-cell stage, but are subsequently repressed (Liu et al., 2019). Notably, DUX4 overexpression in hESCs leads to the binding of DUX4 to 30% of the embryonically accessible MLT2A1 elements (Hendrickson et al., 2017; Liu et al., 2019). DUX4 also activates transcription of various other TEs (Geng et al., 2012; Hendrickson et al., 2017; Vuoristo et al., 2022; Young et al., 2013), and for example, the EGA genes ZSCAN4 and KHDC1P1 are regulated by DUX4-activated ERVL-MaLR overlapping enhancers in DUX4-expressing hESCs (Vuoristo et al., 2022). Some TEs have been evolutionarily co-opted to regulate species- and context-specific gene expression programs (Franke et al., 2017; Hashimoto et al., 2021; Liang et al., 2010; Macaulay et al., 2011; Pontis et al., 2019; Whiddon et al., 2017), and we speculate that some DUX4-induced TEs may have been co-opted to regulate human EGA transcripts.
Mechanisms of DUX4-mediated gene regulation
DUX4 has the capacity to modulate nucleosome structure by inducing the expression and incorporation of histone variants H3.X and H3.Y, which are associated with a relaxed chromatin state and enhanced transcription of the DUX4 target genes (Resnick et al., 2019). In myoblasts, induced DUX4 expression leads to the incorporation of H3.X/Y into highly transcribed DUX4 target genes, potentially contributing to the maintenance of an open chromatin conformation (Resnick et al., 2019). Studies on protein-protein interactions in myoblasts and HEK293 cells have shown that DUX4 can interact with histone acetyltransferases p300 and the CREB binding protein (CBP) (Choi et al., 2016; Vuoristo et al., 2022). These histone acetyltransferases are typically recruited to enhancers by transcription factors, resulting in local acetylation of H3K27 and subsequent expression of target genes (Raisner et al., 2018). DUX4 interacts with p300/CBP through its C-terminus, and the deletion of the last 98 amino acids of the DUX4 C-terminus disrupts the interaction with both p300 and CBP (Choi et al., 2016). The expression of several DUX4 target genes is reduced when the interaction of p300/CBP with DUX4 is disrupted by the DUX4 C-terminal deletion (Choi et al., 2016) or by p300/CBP inhibition (Bosnakovski et al., 2019), suggesting that transcriptional activation of certain DUX4 target genes depends on p300/CBP. Furthermore, DUX4 can interact with several chromatin modifiers and transcriptional modifiers both stably and transiently (Vuoristo et al., 2022). DUX4 interacts with several mediator complex family members that relay regulatory signals from transcription factors to RNA polymerase II (Chen et al., 2021). DUX4 binds MED15 via the six amino acid KIX binding motif, which is located at the very end of the DUX4 C-terminus (Vuoristo et al., 2022). This interaction presents another direct mechanism of DUX4-mediated transcriptional modulation. Besides transcriptional regulation, DUX4 has been implicated in translational regulation. A recent study showed that induced DUX4 expression leads to translational suppression of numerous mRNAs in myoblasts (Hamm et al., 2023). The DUX4-mediated regulation of translation is likely attributed to alterations in the activity of translation initiation regulators 4EBP1, eIF4E, and elongation factor eEF2, however, the intermediate mechanisms responsible for perturbing these factors remain unknown (Hamm et al., 2023). Taken together, DUX4 emerges as a versatile regulator, engaging with a variety of proteins to shape transcription and translation processes (summarized in Fig. 2). Further research is needed to elaborate implications of the DUX4 protein interactions and unravel the precise regulatory mechanisms downstream of DUX4.
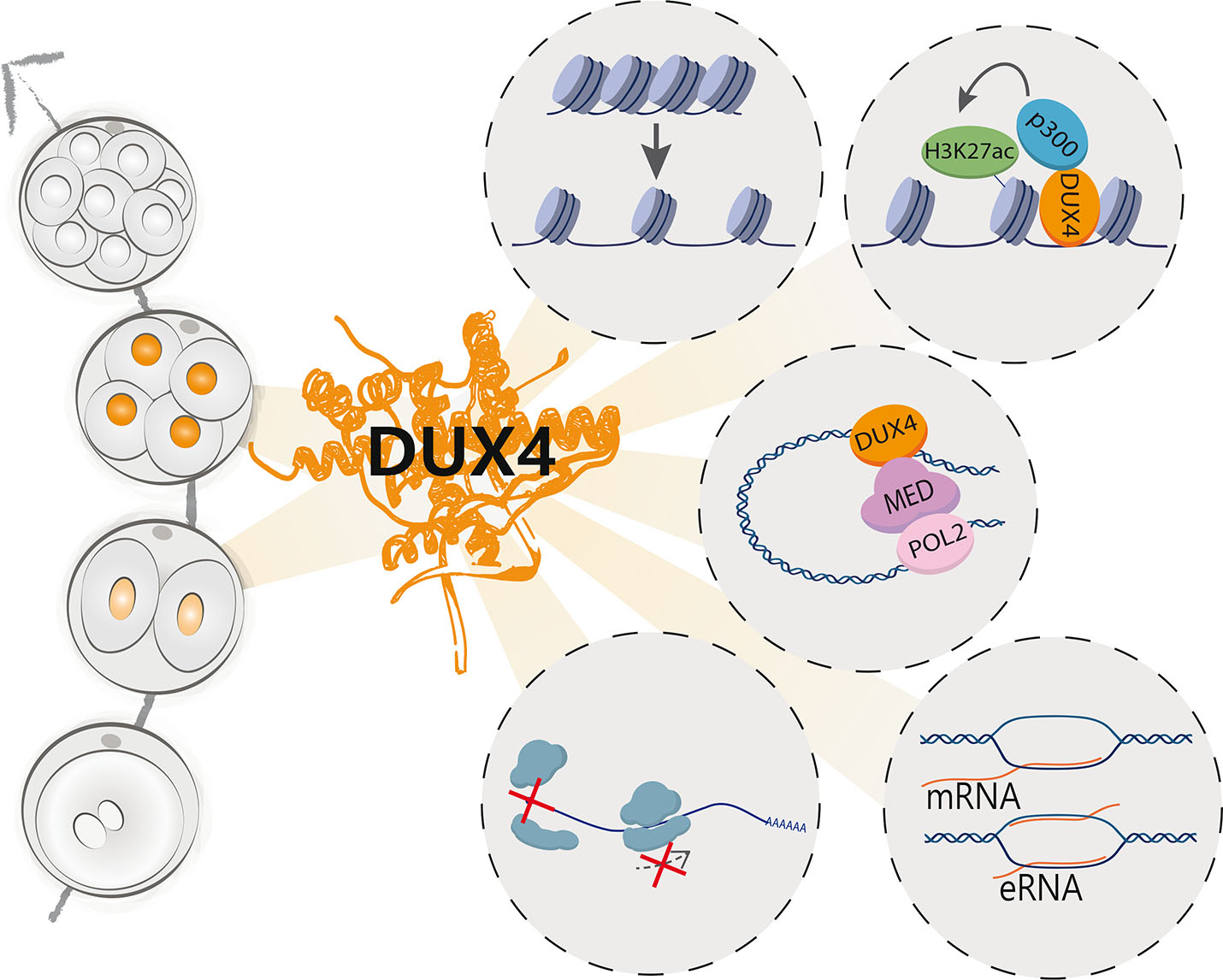
Fig. 2. A summary of the suggested mechanisms of action of DUX4.
DUX4 accumulates in the nuclei of two- and four-cell stage human embryos and is subsequently cleared by the 8-cell stage (left) (Hendrickson et al., 2017; Vuoristo et al., 2022). DUX4 regulates chromatin accessibility (Resnick et al., 2019; Vuoristo et al., 2022), and interacts with various proteins, such as histone acetyltransferase p300 (Choi et al., 2016) and mediator complex members (Vuoristo et al., 2022), which are involved in transcriptional regulation. DUX4 activates several embryonic genome activation (EGA)-associated genes, repetitive elements (De Iaco et al., 2017; Geng et al., 2012; Hendrickson et al., 2017; Liu et al., 2019; Vuoristo et al., 2022) and putative enhancers (Vuoristo et al., 2022), and it was recently implicated in inhibition of translation initiation and elongation (Hamm et al., 2023). Abbreviations: DUX4, double homeobox 4; eRNA, enhancer RNA; H3K27ac, histone 3 lysine 27 acetylation; MED, mediator complex, mRNA, messenger RNA.
Regulation of DUX4 expression
DUX4 transcripts have not been detected in oocytes, indicating that DUX4 is embryonically expressed, however factors and processes responsible for triggering DUX4 expression in human embryos remain unknown. In a study by Grow et al., an LTR element (LTR10C) near the DUX4 gene was identified as a DUX4 enhancer in myoblasts and induced pluripotent stem cells (iPSC) that were derived from FSHD patients (Grow et al., 2021). This enhancer was found to be bound by p53, a transcription factor that is activated in response to DNA damage, and as shown by CRISPR interference is required for full p53-dependent activation of the DUX4 locus in FSHD iPSC (Grow et al., 2021). However, p53 binding alone is insufficient to cause activation of DUX4 expression as the enhancer also becomes occupied by p53 in non-FSHD cells upon induction of DNA damage. Therefore, it is likely that p53-induced DUX4 expression entails additional prerequisites such as inefficient epigenetic repression, as is observed in FSHD (Grow et al., 2021). Notably, telomere shortening, a phenomenon observed both in FSHD (Stadler et al., 2013) and human embryos up to the 4-cell stage, may result in the loss of H3K9 methylated heterochromatin at the DUX4 locus (Zhang et al., 2023). This decrease in epigenetic repression is suggested to facilitate p53 binding to the LTR10C enhancer subsequently leading to the activation of DUX4 (Zhang et al., 2023).
EGA is required for zygotic development, but it is equally important that this unique transcription program is timely repressed as development progresses. The level of H3K9me3, which is strongly associated with densely packed and transcriptionally silenced heterochromatin, gradually accumulates during human pre-implantation development (van de Werken et al., 2014; Xia et al., 2019; Xu et al., 2022; Yu et al., 2022). While some TEs are marked by H3K9me3 throughout development, others gain H3K9me3 in a stage-specific manner (Xu et al., 2022; Yu et al., 2022), strongly implying their developmental stage-specific involvement in cis-regulatory functions during human embryogenesis. Mechanisms behind the selective temporal repression of specific genomic loci during human EGA remains poorly understood. Mouse DUX, a homolog of DUX4, has been suggested to contribute to the establishment of H3K9me3 by inducing the expression of DUXBL, which is subsequently recruited to DUX-bound regions with the TRIM24/33 complex to facilitate the silencing of the associated genes and TEs (Vega-Sendino et al., 2024). A recent study proposes an analogous mechanism in humans, where DUXA that is expressed in the 8-cell stage embryos (Töhönen et al., 2015), potentially contributes to the repression of DUX4 target loci (Bosnakovski et al., 2023). DUXA DNA binding motifs are highly similar to those of DUX4 (Liu et al., 2019), however DUXA lacks the C-terminal transactivating domain, presumably abolishing its ability to activate transcription (Bosnakovski et al., 2023). Remarkably, when both DUX4 and DUXA are ectopically expressed in myoblasts, the expression levels of DUX4 target genes are significantly reduced (Bosnakovski et al., 2023). DUXA expression is activated by DUX4, which points to a feedback inhibition mechanism where DUXA suppresses DUX4 targets, possibly as a result of competitive target sequence binding by DUX4 and DUXA (Bosnakovski et al., 2023).
The mechanisms of DUX4 repression in human embryos and somatic tissues remain unclear, however it is conceivable that multiple epigenetic mechanisms, such as DNA methylation and H3K9me3 mediated heterochromatin formation, contribute to this. In mouse 2-cell embryos, the relocation of Dux loci to the nucleolar periphery potentially drives the repression of Dux through the formation of perinucleolar heterochromatin, concurrently with the maturation of nucleoli (Xie et al., 2022; Yu et al., 2021). Also in human embryos, nucleoli maturation initiates around the time of EGA (Tesarik et al., 1986) and coincides with the repression of DUX4 (Kresoja-Rakic and Santoro, 2019). However, whether nucleolar maturation contributes to DUX4 repression in humans remains a topic for future research. Due to the location of DUX4 in the subtelomeric region, it may be affected by the transcriptional repression induced by the spread of telomeric heterochromatin, named telomere position effect (TPE) (Lee et al., 2021). Zhang et al., have provided evidence that telomere extension may contribute to the silencing of DUX4 in human embryos, supported by the observation that telomere length extends during major ZGA (Zhang et al., 2023) and longer telomeres lead to pronounced TPE (Baur et al., 2001).
Future prospects
Ever since DUX4 was recognized as a potent EGA factor it has raised broad interest across the research community. Earlier findings about DUX4 that were mainly focused on the etiology of FSHD, have become augmented by recent perceptions that DUX4 actively modulates embryonic gene expression and embryonic development. Yet several important questions about DUX4 and its implications in human oocyte-to-embryo transition and EGA remain unanswered. Current DUX4 annotations rely on the DUX4 sequences found in human somatic cells like myoblasts and therefore, it is possible that human zygotes express DUX4 sequence variants. Cloning and sequencing of the full-length DUX4 mRNA from zygotes and cleavage stage embryos using traditional PCR-based technologies may be complicated due to the repetitive nature and high GC content of this transcript (Jagannathan et al., 2016). Therefore, recent technological improvements in long read sequencing may prove to be useful in determining the DUX4 mRNA sequence in human embryonic samples. Relatedly, recent findings according to which the canonical DUX4 interacts with numerous chromatin modifiers, RNA-binding proteins, and transcriptional modifiers open an interesting avenue to study which of these interactions take place in a context-dependent manner and how these interactions may pertain to embryonic development.
Our understanding about the implications of DUX4 in human development is restricted, mainly due to ethical and technical limitations related to experiments where human embryos are used. Downscaling the amount of input material needed to perform for instance chromatin profiling has enabled informative analyses about oocytes and preimplantation human embryos. As an example, LiCAT-sequencing that requires low input material (Liu et al., 2019) allowed Zhang et al., to correlate chromatin accessibility at the DUX4 regulatory region with DUX4 expression levels and telomere length during EGA (Zhang et al., 2023). These types of approaches clarify the possible mechanisms behind the onset of DUX4 expression in human development. While mutating of the DUX4 transcription start site in human zygotes leads to embryo stalling by the 8-cell stage (Liu et al., 2022), the knockdown of DUX4 leads to impaired oocyte-to-embryo transition (Vuoristo et al., 2022). This indicates that RNAi-mediated knockdown of DUX4 at zygotic stage may not reveal a complete DUX4 phenotype. The EGA-associated coding transcripts are relatively well-known, however another interesting research avenue will be to investigate the implications of TEs that are activated at the time of EGA. Transcriptome and translatome profiling of preimplantation human embryos in combination with extensive functional experiments emphasized the importance of both maternal and zygotic transcription factors (Zou et al., 2022). These factors include TPRXL and OTX2 that are of maternal origin and undergo translation starting from the oocyte meiotic resumption (Zou et al., 2022). The OTX2 is highly expressed in human oocytes (Xue et al., 2013; Yan et al., 2013), and its binding motif is enriched at accessible chromatin regions in the early embryo (Liu et al., 2019), indicating that it functions temporally in parallel with DUX4. Simultaneous knockdown of TPRXL and embryonically expressed TPRX1 and TPRX2 results in developmental delay and impaired EGA, while knockdown of these factors individually caused milder phenotypes (Zou et al., 2022). EGA-associated factors presumably form a transcriptional circuitry in their specific epigenome landscape. Further research is imperative to understand how these factors function and to what extent each of these factors may be indispensable for development or whether they can compensate for one another. One of the challenges related to research on human EGA factors is their highly unique in vivo state, which is difficult to recapitulate in vitro. Recently acknowledged 8-cell embryo-like cellular model systems provide promising platforms for future studies (Mazid et al., 2022; Moya-Jódar et al., 2023; Taubenschmid-Stowers et al., 2022; Yoshihara et al., 2022; Yu et al., 2022), although they lack some of the key in vivo aspects such as the presence of maternal factors. As a summary, recent studies have emphasized the role of DUX4 as one of the active transcription factors during human EGA and focused on discovering how DUX4 may become activated, and how it regulates its target sequences. Future research is imperative to further elucidate mechanisms of DUX4 and other EGA factors in the context of human EGA and in cellular reprogramming.
Acknowledgements
We acknowledge MSc. Pauliina Paloviita, Dr. Michelle Percharde and Dr. Edward Grow for critical reading of the manuscript and helpful comments. This work was supported by the University of Helsinki Doctoral Programme in Biomedicine (S.N.), the Finnish Fertility Society (S.N.), the University of Helsinki Three-year grant (S.V.), the Sigrid Jusélius Senior Researcher grant (S.V.), the Helsinki Institute of Life Science Fellowship (S.V.), and the Academy of Finland Academy fellowship grant #348111 and #353549 (S.V.).
Abbreviations
DUX4, Double Homeobox 4 ; EGA, Embryonic genome activation ; FSHD, Facioscapulohumeral muscular dystrophy ; TE, Transposable element ;References
Almeida M. V., Vernaz G., Putman A. L. K., Miska E. A. (2022). Taming transposable elements in vertebrates: from epigenetic silencing to domestication. Trends in Genetics 38: 529-553.
Baur J. A., Zou Y., Shay J. W., Wright W. E. (2001). Telomere Position Effect in Human Cells. Science 292: 2075-2077.
Bosnakovski D., da Silva M. T., Sunny S. T., Ener E. T., Toso E. A., Yuan C., Cui Z., Walters M. A., Jadhav A., Kyba M. (2019). A novel P300 inhibitor reverses DUX4-mediated global histone H3 hyperacetylation, target gene expression, and cell death. Science Advances 5: eaaw7781.
Bosnakovski D., Lamb S., Simsek T., Xu Z., Belayew A., Perlingeiro R., Kyba M. (2008). DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Experimental Neurology 214: 87-96.
Bosnakovski D., Toso E. A., Ener E. T., Gearhart M. D., Yin L., Lüttmann F. F., Magli A., Shi K., Kim J., Aihara H., Kyba M. (2023). Antagonism among DUX family members evolved from an ancestral toxic single homeodomain protein. iScience 26: 107823.
Bouckenheimer J., Assou S., Riquier S., Hou C., Philippe N., Sansac C., Lavabre-Bertrand T., Commes T., Lemaître J.M., Boureux A., De Vos J. (2016). Long non-coding RNAs in human early embryonic development and their potential in ART. Human Reproduction Update 23: 19-40.
Bourque G., Burns K. H., Gehring M., Gorbunova V., Seluanov A., Hammell M., Imbeault M., Izsvák Z., Levin H. L., Macfarlan T. S., Mager D. L., Feschotte C. (2018). Ten things you should know about transposable elements. Genome Biology 19: 199.
Braude P., Bolton V., Moore S. (1988). Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332: 459-461.
Campbell A. E., Belleville A. E., Resnick R., Shadle S. C., Tapscott S. J. (2018). Facioscapulohumeral dystrophy: activating an early embryonic transcriptional program in human skeletal muscle. Human Molecular Genetics 27: R153-R162.
Campbell A. E., Dyle M. C., Albanese R., Matheny T., Sudheendran K., Cortázar M. A., Forman T., Fu R., Gillen A. E., Caruthers M. H., Floor S. N., Calviello L., Jagannathan S. (2023). Compromised nonsense-mediated RNA decay results in truncated RNA-binding protein production upon DUX4 expression. Cell Reports 42: 112642.
Chen X., Ke Y., Wu K., Zhao H., Sun Y., Gao L., Liu Z., Zhang J., Tao W., Hou Z., Liu H., Liu J., Chen Z.J. (2019). Key role for CTCF in establishing chromatin structure in human embryos. Nature 576: 306-310.
Chen X., Yin X., Li J., Wu Z., Qi Y., Wang X., Liu W., Xu Y. (2021). Structures of the human Mediator and Mediator-bound preinitiation complex. Science 372: 6546.
Choi S. H., Gearhart M. D., Cui Z., Bosnakovski D., Kim M., Schennum N., Kyba M. (2016). DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Research 44: 5161-5173.
De Iaco A., Planet E., Coluccio A., Verp S., Duc J., Trono D. (2017). DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nature Genetics 49: 941-945.
DiRusso J. A., Clark A. T. (2023). Transposable elements in early human embryo development and embryo models. Current Opinion in Genetics & Development 81: 102086.
Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A. M., Leo O., Figlewicz D., Barro M., Laoudj-Chenivesse D., Belayew A., Coppée F., Chen Y.W. (2007). DUX4 , a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1 . Proceedings of the National Academy of Sciences 104: 18157-18162.
Feng Q., Snider L., Jagannathan S., Tawil R., van der Maarel S. M., Tapscott S. J., Bradley R. K. (2015). A feedback loop between nonsense-mediated decay and the retrogene DUX4 in facioscapulohumeral muscular dystrophy. eLife 4: e04996.
Ferrie J. J., Karr J. P., Tjian R., Darzacq X. (2022). “Structure”-function relationships in eukaryotic transcription factors: The role of intrinsically disordered regions in gene regulation. Molecular Cell 82: 3970-3984.
Franke V., Ganesh S., Karlic R., Malik R., Pasulka J., Horvat F., Kuzman M., Fulka H., Cernohorska M., Urbanova J., Svobodova E., Ma J., Suzuki Y., Aoki F., Schultz R. M., Vlahovicek K., Svoboda P. (2017). Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Research 27: 1384-1394.
Gabriëls J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., Belayew A. (1999). Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene 236: 25-32.
Gainetdinov I., Skvortsova Y., Kondratieva S., Funikov S., Azhikina T. (2017). Two modes of targeting transposable elements by piRNA pathway in human testis. RNA 23: 1614-1625.
Gao L., Wu K., Liu Z., Yao X., Yuan S., Tao W., Yi L., Yu G., Hou Z., Fan D., Tian Y., Liu J., Chen Z.J., Liu J. (2018). Chromatin Accessibility Landscape in Human Early Embryos and Its Association with Evolution. Cell 173: 248-259.e15.
Gawriyski L., Jouhilahti E.M., Yoshihara M., Fei L., Weltner J., Airenne T. T., Trokovic R., Bhagat S., Tervaniemi M. H., Murakawa Y., Salokas K., Liu X., Miettinen S., Bürglin T. R., Sahu B., Otonkoski T., Johnson M. S., Katayama S., Varjosalo M., Kere J. (2023). Comprehensive characterization of the embryonic factor LEUTX. iScience 26: 106172.
Geng L. N., Yao Z., Snider L., Fong A. P., Cech J. N., Young J. M., van der Maarel S. M., Ruzzo W. L., Gentleman R. C., Tawil R., Tapscott S. J. (2012). DUX4 Activates Germline Genes, Retroelements, and Immune Mediators: Implications for Facioscapulohumeral Dystrophy. Developmental Cell 22: 38-51.
Göke J., Lu X., Chan Y.S., Ng H.H., Ly L.H., Sachs F., Szczerbinska I. (2015). Dynamic Transcription of Distinct Classes of Endogenous Retroviral Elements Marks Specific Populations of Early Human Embryonic Cells. Cell Stem Cell 16: 135-141.
Grow E. J., Flynn R. A., Chavez S. L., Bayless N. L., Wossidlo M., Wesche D. J., Martin L., Ware C. B., Blish C. A., Chang H. Y., Reijo Pera R. A., Wysocka J. (2015). Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522: 221-225.
Grow E. J., Weaver B. D., Smith C. M., Guo J., Stein P., Shadle S. C., Hendrickson P. G., Johnson N. E., Butterfield R. J., Menafra R., Kloet S. L., van der Maarel S. M., Williams C. J., Cairns B. R. (2021). p53 convergently activates Dux/DUX4 in embryonic stem cells and in facioscapulohumeral muscular dystrophy cell models. Nature Genetics 53: 1207-1220.
Hamm D. C., Paatela E. M., Bennett S. R., Wong C.J., Campbell A. E., Wladyka C. L., Smith A. A., Jagannathan S., Hsieh A. C., Tapscott S. J. (2023). The transcription factor DUX4 orchestrates translational reprogramming by broadly suppressing translation efficiency and promoting expression of DUX4-induced mRNAs. PLOS Biology 21: e3002317.
Hashimoto K., Jouhilahti E.M., Töhönen V., Carninci P., Kere J., Katayama S. (2021). Embryonic LTR retrotransposons supply promoter modules to somatic tissues. Genome Research 31: 1983-1993.
Hendrickson P. G., Doráis J. A., Grow E. J., Whiddon J. L., Lim J.W., Wike C. L., Weaver B. D., Pflueger C., Emery B. R., Wilcox A. L., Nix D. A., Peterson C. M., Tapscott S. J., Carrell D. T., Cairns B. R. (2017). Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nature Genetics 49: 925-934.
Hewitt J. E. (2015). Loss of epigenetic silencing of the DUX4 transcription factor gene in facioscapulohumeral muscular dystrophy: Figure 1.. Human Molecular Genetics 24: R17-R23.
Jagannathan S., Ogata Y., Gafken P. R., Tapscott S. J., Bradley R. K. (2019). Quantitative proteomics reveals key roles for post-transcriptional gene regulation in the molecular pathology of facioscapulohumeral muscular dystrophy. eLife 8: e41740.
Jagannathan S., Shadle S. C., Resnick R., Snider L., Tawil R. N., van der Maarel S. M., Bradley R. K., Tapscott S. J. (2016). Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Human Molecular Genetics 25: 4419-4431.
Janssen A., Colmenares S. U., Karpen G. H. (2018). Heterochromatin: Guardian of the Genome. Annual Review of Cell and Developmental Biology 34: 265-288.
Kowaljow V., Marcowycz A., Ansseau E., Conde C. B., Sauvage S., Mattéotti C., Arias C., Corona E. D., Nuñez N. G., Leo O., Wattiez R., Figlewicz D., Laoudj-Chenivesse D., Belayew A., Coppée F., Rosa A. L. (2007). The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscular Disorders 17: 611-623.
Kresoja-Rakic J., Santoro R. (2019). Nucleolus and rRNA Gene Chromatin in Early Embryo Development. Trends in Genetics 35: 868-879.
Lee J. K., Bosnakovski D., Toso E. A., Dinh T., Banerjee S., Bohl T. E., Shi K., Orellana K., Kyba M., Aihara H. (2018). Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA. Cell Reports 25: 2955-2962.e3.
Lee K.H., Kim D.Y., Kim W. (2021). Regulation of Gene Expression by Telomere Position Effect. International Journal of Molecular Sciences 22: 12807.
Leidenroth A., Clapp J., Mitchell L. M., Coneyworth D., Dearden F. L., Iannuzzi L., Hewitt J. E. (2012). Evolution of DUX gene macrosatellites in placental mammals. Chromosoma 121: 489-497.
Leidenroth A., Hewitt J. E. (2010). A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evolutionary Biology 10: 364.
Lemmers R. J. L. F., van der Vliet P. J., Klooster R., Sacconi S., Camaño P., Dauwerse J. G., Snider L., Straasheijm K. R., Jan van Ommen G., Padberg G. W., Miller D. G., Tapscott S. J., Tawil R., Frants R. R., van der Maarel S. M. (2010). A Unifying Genetic Model for Facioscapulohumeral Muscular Dystrophy. Science 329: 1650-1653.
Liang C.Y., Wang L.J., Chen C.P., Chen L.F., Chen Y.H., Chen H. (2010). GCM1 Regulation of the Expression of Syncytin 2 and Its Cognate Receptor MFSD2A in Human Placenta1. Biology of Reproduction 83: 387-395.
Liu L., Leng L., Liu C., Lu C., Yuan Y., Wu L., Gong F., Zhang S., Wei X., Wang M., Zhao L., Hu L., Wang J., Yang H., Zhu S., Chen F., Lu G., Shang Z., Lin G. (2019). An integrated chromatin accessibility and transcriptome landscape of human pre-implantation embryos. Nature Communications 10: 364.
Liu Y., Lu X., Ye M., Wang L., Tang R., Yang Z., Turathum B., Liu C., Xue Y., Wu M., Yang Y., Gao E., Zhang D., Yang F., Kee K.K., Huang X., Li G., Chian R.C. (2022). Efficient silencing of the multicopy DUX4 gene by ABE-mediated start codon mutation in human embryos. Journal of Genetics and Genomics 49: 982-985.
Macaulay E. C., Weeks R. J., Andrews S., Morison I. M. (2011). Hypomethylation of functional retrotransposon-derived genes in the human placenta. Mammalian Genome 22: 722-735.
Madissoon E., Jouhilahti E.M., Vesterlund L., Töhönen V., Krjutškov K., Petropoulos S., Einarsdottir E., Linnarsson S., Lanner F., Månsson R., Hovatta O., Bürglin T. R., Katayama S., Kere J. (2016). Characterization and target genes of nine human PRD-like homeobox domain genes expressed exclusively in early embryos. Scientific Reports 6: 28995.
Mazid M. A., Ward C., Luo Z., Liu C., Li Y., Lai Y., Wu L., Li J., Jia W., Jiang Y., Liu H., Fu L., Yang Y., Ibañez D. P., Lai J., Wei X., An J., Guo P., Yuan Y., Deng Q., Wang Y., Liu Y., Gao F., Wang J., Zaman S., Qin B., Wu G., Maxwell P. H., Xu X., Liu L., Li W., Esteban M. A. (2022). Rolling back human pluripotent stem cells to an eight-cell embryo-like stage. Nature 605: 315-324.
Mitsuhashi H., Ishimaru S., Homma S., Yu B., Honma Y., Beermann M. L., Miller J. B. (2018). Functional domains of the FSHD-associated DUX4 protein. Biology Open 7: bio033977.
Moya-Jódar M., Ullate-Agote A., Barlabé P., Rodríguez-Madoz J. R., Abizanda G., Barreda C., Carvajal-Vergara X., Vilas-Zornoza A., Romero J. P., Garate L., Agirre X., Coppiello G., Prósper F., Aranguren X. L. (2023). Revealing cell populations catching the early stages of human embryo development in naive pluripotent stem cell cultures. Stem Cell Reports 18: 64-80.
Paloviita P., Vuoristo S. (2022). The non-coding genome in early human development – Recent advancements. Seminars in Cell & Developmental Biology 131: 4-13.
Petropoulos S., Panula S. P., Schell J. P., Lanner F. (2016). Single‐cell RNA sequencing: revealing human pre‐implantation development, pluripotency and germline development . Journal of Internal Medicine 280: 252-264.
Pontis J., Planet E., Offner S., Turelli P., Duc J., Coudray A., Theunissen T. W., Jaenisch R., Trono D. (2019). Hominoid-Specific Transposable Elements and KZFPs Facilitate Human Embryonic Genome Activation and Control Transcription in Naive Human ESCs. Cell Stem Cell 24: 724-735.e5.
Raisner R., Kharbanda S., Jin L., Jeng E., Chan E., Merchant M., Haverty P. M., Bainer R., Cheung T., Arnott D., Flynn E. M., Romero F. A., Magnuson S., Gascoigne K. E. (2018). Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Reports 24: 1722-1729.
Resnick R., Wong C.J., Hamm D. C., Bennett S. R., Skene P. J., Hake S. B., Henikoff S., van der Maarel S. M., Tapscott S. J. (2019). DUX4-Induced Histone Variants H3.X and H3.Y Mark DUX4 Target Genes for Expression. Cell Reports 29: 1812-1820.e5.
Rickard A. M., Petek L. M., Miller D. G. (2015). Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Human Molecular Genetics 24: 5901-5914.
Sacconi S., Briand-Suleau A., Gros M., Baudoin C., Lemmers R. J.L.F., Rondeau S., Lagha N., Nigumann P., Cambieri C., Puma A., Chapon F., Stojkovic T., Vial C., Bouhour F., Cao M., Pegoraro E., Petiot P., Behin A., Marc B., Eymard B., Echaniz-Laguna A., Laforet P., Salviati L., Jeanpierre M., Cristofari G., van der Maarel S. M. (2019). FSHD1 and FSHD2 form a disease continuum. Neurology 92: e2273>-e2285.
Schulz K. N., Harrison M. M. (2019). Mechanisms regulating zygotic genome activation. Nature Reviews Genetics 20: 221-234.
Shadle S. C., Zhong J. W., Campbell A. E., Conerly M. L., Jagannathan S., Wong C.J., Morello T. D., van der Maarel S. M., Tapscott S. J. (2017). DUX4-induced dsRNA and MYC mRNA stabilization activate apoptotic pathways in human cell models of facioscapulohumeral dystrophy. PLOS Genetics 13: e1006658.
Smith A. A., Nip Y., Bennett S. R., Hamm D. C., Lemmers R. J.L.F., van der Vliet P. J., Setty M., van der Maarel S. M., Tapscott S. J. (2023). DUX4 expression in cancer induces a metastable early embryonic totipotent program. Cell Reports 42: 113114.
Snider L., Geng L. N., Lemmers R. J. L. F., Kyba M., Ware C. B., Nelson A. M., Tawil R., Filippova G. N., van der Maarel S. M., Tapscott S. J., Miller D. G. (2010). Facioscapulohumeral Dystrophy: Incomplete Suppression of a Retrotransposed Gene. PLoS Genetics 6: e1001181.
Stadler G., Rahimov F., King O. D., Chen J. C. J., Robin J. D., Wagner K. R., Shay J. W., Emerson C. P., Wright W. E. (2013). Telomere position effect regulates DUX4 in human facioscapulohumeral muscular dystrophy. Nature Structural & Molecular Biology 20: 671-678.
Taubenschmid-Stowers J., Rostovskaya M., Santos F., Ljung S., Argelaguet R., Krueger F., Nichols J., Reik W. (2022). 8C-like cells capture the human zygotic genome activation program in vitro. Cell Stem Cell 29: 449-459.e6.
Tesařék J., Kopečný V., Plachot M., Mandelbaum J. (1988). Early morphological signs of embryonic genome expression in human preimplantation development as revealed by quantitative electron microscopy. Developmental Biology 128: 15-20.
Tesarik J., Kopecny V., Plachot M., Mandelbaum J. (1986). Activation of nucleolar and extranucleolar RNA synthesis and changes in the ribosomal content of human embryos developing in vitro. Reproduction 78: 463-470.
Töhönen V., Katayama S., Vesterlund L., Jouhilahti E.M., Sheikhi M., Madissoon E., Filippini-Cattaneo G., Jaconi M., Johnsson A., Bürglin T. R., Linnarsson S., Hovatta O., Kere J. (2015). Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nature Communications 6: 8207.
Töhönen V., Katayama S., Vesterlund L., Sheikhi M., Antonsson L., Filippini-Cattaneo G., Jaconi M., Johnsson A., Linnarsson S., Hovatta O., Kere J., (2017). Transcription activation of early human development suggests DUX4 as an embryonic regulator. bioRxiv Preprint: 123208.
van de Werken C., van der Heijden G. W., Eleveld C., Teeuwssen M., Albert M., Baarends W. M., Laven J. S. E., Peters A. H. F. M., Baart E. B. (2014). Paternal heterochromatin formation in human embryos is H3K9/HP1 directed and primed by sperm-derived histone modifications. Nature Communications 5: 5868.
Vassena R., Boué S., González-Roca E., Aran B., Auer H., Veiga A., Belmonte J. C. I. (2011). Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138: 3699-3709.
Vastenhouw N. L., Cao W. X., Lipshitz H. D. (2019). The maternal-to-zygotic transition revisited. Development 146: dev161471.
Vega-Sendino M., Lüttmann F. F., Olbrich T., Chen Y., Kuenne C., Stein P., Tillo D., Carey G. I., Zhong J., Savy V., Radonova L., Lu T., Saykali B., Kim K. P., Domingo C. N., Schüler L., Günther S., Bentsen M., Bosnakovski D., Schöler H., Kyba M., Maity T. K., Jenkins L. M., Looso M., Williams C. J., Kim J., Ruiz S., (2024). The homeobox transcription factor DUXBL controls exit from totipotency. Nature genetics 56: 697-709.
Vuoristo S., Bhagat S., Hydén-Granskog C., Yoshihara M., Gawriyski L., Jouhilahti E.M., Ranga V., Tamirat M., Huhtala M., Kirjanov I., Nykänen S., Krjutškov K., Damdimopoulos A., Weltner J., Hashimoto K., Recher G., Ezer S., Paluoja P., Paloviita P., Takegami Y., Kanemaru A., Lundin K., Airenne T. T., Otonkoski T., Tapanainen J. S., Kawaji H., Murakawa Y., Bürglin T. R., Varjosalo M., Johnson M. S., Tuuri T., Katayama S., Kere J. (2022). DUX4 is a multifunctional factor priming human embryonic genome activation. iScience 25: 104137.
Wallace L. M., Garwick S. E., Mei W., Belayew A., Coppee F., Ladner K. J., Guttridge D., Yang J., Harper S. Q. (2011). DUX4 , a candidate gene for facioscapulohumeral muscular dystrophy, causes p53‐dependent myopathy in vivo . Annals of Neurology 69: 540-552.
Whiddon J. L., Langford A. T., Wong C.J., Zhong J. W., Tapscott S. J. (2017). Conservation and innovation in the DUX4-family gene network. Nature Genetics 49: 935-940.
Wu J., Xu J., Liu B., Yao G., Wang P., Lin Z., Huang B., Wang X., Li T., Shi S., Zhang N., Duan F., Ming J., Zhang X., Niu W., Song W., Jin H., Guo Y., Dai S., Hu L., Fang L., Wang Q., Li Y., Li W., Na J., Xie W., Sun Y. (2018). Chromatin analysis in human early development reveals epigenetic transition during ZGA. Nature 557: 256-260.
Xia W., Xu J., Yu G., Yao G., Xu K., Ma X., Zhang N., Liu B., Li T., Lin Z., Chen X., Li L., Wang Q., Shi D., Shi S., Zhang Y., Song W., Jin H., Hu L., Bu Z., Wang Y., Na J., Xie W., Sun Y.P. (2019). Resetting histone modifications during human parental-to-zygotic transition. Science 365: 353-360.
Xie S. Q., Leeke B. J., Whilding C., Wagner R. T., Garcia-Llagostera F., Low Y.X., Chammas P., Cheung N. T.F., Dormann D., McManus M. T., Percharde M. (2022). Nucleolar-based Dux repression is essential for embryonic two-cell stage exit . Genes & Development 36: 331-347.
Xu R., Li S., Wu Q., Li C., Jiang M., Guo L., Chen M., Yang L., Dong X., Wang H., Wang C., Liu X., Ou X., Gao S. (2022). Stage-specific H3K9me3 occupancy ensures retrotransposon silencing in human pre-implantation embryos. Cell Stem Cell 29: 1051-1066.e8.
Xue Z., Huang K., Cai C., Cai L., Jiang C., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y. E., Liu J., Horvath S., Fan G. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500: 593-597.
Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., Huang J., Li M., Wu X., Wen L., Lao K., Li R., Qiao J., Tang F. (2013). Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nature Structural & Molecular Biology 20: 1131-1139.
Yoshihara M., Kirjanov I., Nykänen S., Sokka J., Weltner J., Lundin K., Gawriyski L., Jouhilahti E.M., Varjosalo M., Tervaniemi M. H., Otonkoski T., Trokovic R., Katayama S., Vuoristo S., Kere J. (2022). Transient DUX4 expression in human embryonic stem cells induces blastomere-like expression program that is marked by SLC34A2. Stem Cell Reports 17: 1743-1756.
Young J. M., Whiddon J. L., Yao Z., Kasinathan B., Snider L., Geng L. N., Balog J., Tawil R., van der Maarel S. M., Tapscott S. J. (2013). DUX4 Binding to Retroelements Creates Promoters That Are Active in FSHD Muscle and Testis. PLoS Genetics 9: e1003947.
Yu H., Chen M., Hu Y., Ou S., Yu X., Liang S., Li N., Yang M., Kong X., Sun C., Jia S., Zhang Q., Liu L., Hurst L. D., Li R., Wang W., Wang J. (2022). Dynamic reprogramming of H3K9me3 at hominoid-specific retrotransposons during human preimplantation development. Cell Stem Cell 29: 1031-1050.e12.
Yu H., Sun Z., Tan T., Pan H., Zhao J., Zhang L., Chen J., Lei A., Zhu Y., Chen L., Xu Y., Liu Y., Chen M., Sheng J., Xu Z., Qian P., Li C., Gao S., Daley G. Q., Zhang J. (2021). rRNA biogenesis regulates mouse 2C-like state by 3D structure reorganization of peri-nucleolar heterochromatin. Nature Communications 12: 6365.
Yu X., Liang S., Chen M., Yu H., Li R., Qu Y., Kong X., Guo R., Zheng R., Izsvák Z., Sun C., Yang M., Wang J. (2022). Recapitulating early human development with 8C-like cells. Cell Reports 39: 110994.
Zhang X., Zhang C., Zhou D., Zhang T., Chen X., Ren J., He C., Meng F., Zhou Q., Yang Q., Dai C., Lin G., Zeng S., Leng L. (2023). Telomeres cooperate in zygotic genome activation by affecting DUX4/Dux transcription. iScience 26: 106158.
Zou Z., Zhang C., Wang Q., Hou Z., Xiong Z., Kong F., Wang Q., Song J., Liu B., Liu B., Wang L., Lai F., Fan Q., Tao W., Zhao S., Ma X., Li M., Wu K., Zhao H., Chen Z.J., Xie W. (2022). Translatome and transcriptome co-profiling reveals a role of TPRXs in human zygotic genome activation. Science 378: abo7923.