Int. J. Dev. Biol. 68: 169 - 188 (2024)
Special Issue: Developmental Biology in Nordic Countries
Enhancer-promoter communication in Drosophila developmental gene transcription
Open Access | Review | Published: 3 June 2024
Abstract
Enhancers play an essential role in gene regulation by receiving cues from transcription factors and relaying these signals to modulate transcription from target promoters. Enhancer-promoter communications occur across large linear distances of the genome and with high specificity. The molecular mechanisms that underlie enhancer-mediated control of transcription remain unresolved. In this review, we focus on research in Drosophila uncovering the molecular mechanisms governing enhancer-promoter communication and discuss the current understanding of developmental gene regulation. The functions of protein acetylation, pausing of RNA polymerase II, transcriptional bursting, and the formation of nuclear hubs in the induction of tissue-specific programs of transcription during zygotic genome activation are considered.
Keywords
enhancer, transcription, gene regulation, Pol II pausing, transcriptional bursting
Introduction
The ability to express different complements of genes underlies the capacity of cells carrying the same DNA genome to form diverse cell types during multicellular development. Transcription fundamentally depends on recruitment of the RNA polymerase II (Pol II) transcriptional machinery at promoter sequences, but regulation by non-coding enhancer sequences shape the magnitude and spatiotemporal dynamics of transcriptional activity (Shlyueva et al., 2014; Spitz and Furlong, 2012). Enhancers are essential cis-regulatory elements (CREs) that integrate regulatory signals from trans-acting transcription factors (TFs) into a transcriptional circuitry to drive cell type-specific programs of transcription from target promoters (Fig. 1). Enhancers are often located at large genomic distances from target promoters and must therefore communicate regulatory signals across the chromatin landscape that houses the DNA genome (Furlong and Levine, 2018). The genome is organized across multiple levels with chromosomes occupying territories within the nucleus, the segregation of chromatin into active and inactive compartments, topologically associating domains (TADs) with abundant internal interactions, including between enhancers and promoters (Jerkovic and Cavalli, 2021). Cofactors with enzymatic chromatin-modifying activities are often recruited by TFs to enhancers (Reiter et al., 2017) and it is increasingly recognized that modulation of chromatin, through histone posttranslational modifications (PTMs), the incorporation of histone variants, and DNA methylation, interplay with transcriptional states to ensure developmental gene regulation (Bannister and Kouzarides, 2011; Li et al., 2007). While enhancers are well defined as recruitment platforms for cell type-specific TFs with dense enrichment of TF binding sites (TFBS) (Shlyueva et al., 2014; Spitz and Furlong, 2012), mechanistic understanding of how enhancers communicate regulatory cues to promoters and modulate transcriptional activity is lacking (Catarino and Stark, 2018; Furlong and Levine, 2018; Karr et al., 2022; Panigrahi and O'Malley, 2021).
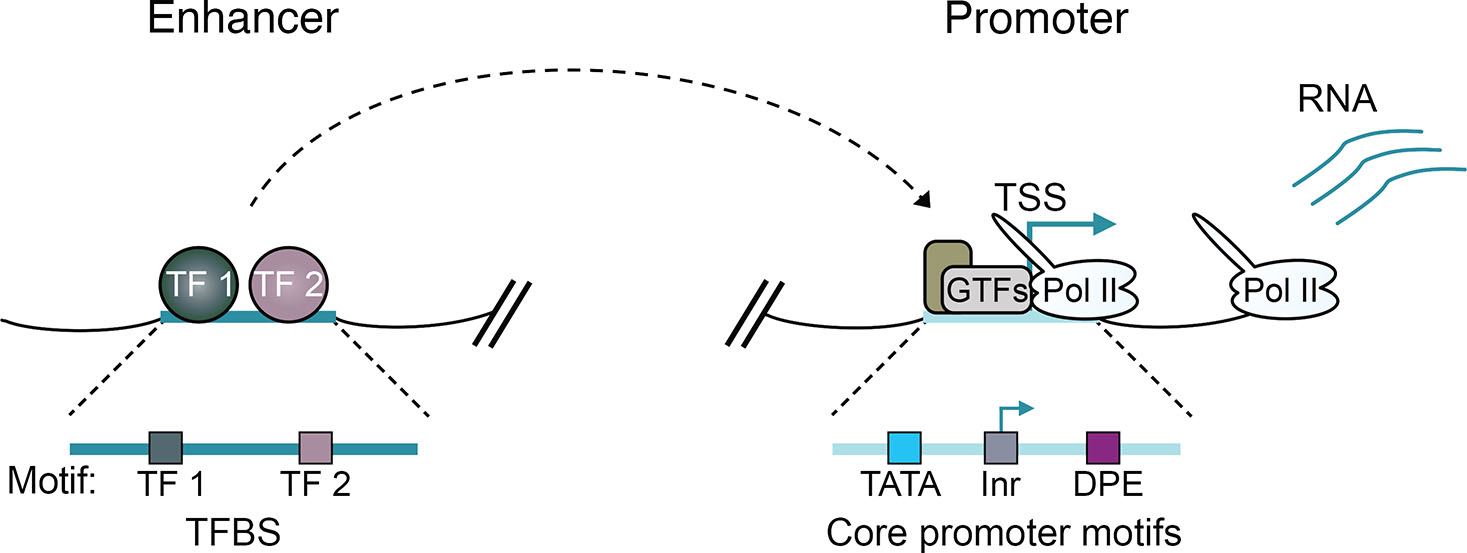
Fig. 1. Enhancers and promoters integrate regulatory signals from transcription factors (TFs) to the genome to drive transcription from promoters.
The schematic depicts the core circuitry of enhancers and promoters, highlighting transcription factor binding sites (TFBS) that recruit TFs to enhancers. Additionally, core promoter motifs, such as the TATA box, initiator element (Inr), and downstream promoter element (DPE), are shown. These motifs play a crucial role in recruiting general transcription factors (GTFs) and the RNA Polymerase II (Pol II) machinery to promoters, to initiate the transcription process.
Drosophila melanogaster (Drosophila) is a key model system for studying the molecular mechanisms underlying developmental gene regulation. During the first few hours of Drosophila embryogenesis, development progresses rapidly from a transcriptionally inert fertilized egg, through a series of 13 synchronous syncytial nuclear cycles (nc), to an embryo composed of ~6000 cells that undergoes zygotic genome activation (ZGA) and initiates cell type-specific transcriptional programs (Harrison and Eisen, 2015; Schulz and Harrison, 2019). ZGA is orchestrated by maternally supplied mRNAs and proteins which establish an elaborate gene regulatory landscape. Chromatin becomes accessible at specific CREs and differentially bound across cells by complements of TFs, cofactors and the transcriptional machinery, while chromatin states, with specific signatures of PTMs, form (Blythe and Wieschaus, 2016; Bozek et al., 2019; Calderon et al., 2022; Cusanovich et al., 2018; Koenecke et al., 2016; Li et al., 2014; Liang et al., 2008; Reddington et al., 2020). ZGA occurs concomitantly with formation of a complex genome organization of TADs that may facilitate close proximity between enhancers and promoters. However, the role of TADs in gene expression and the mechanisms of enhancer-promoter interactions are unclear (Furlong and Levine, 2018; Ghavi-Helm et al., 2014; Hug et al., 2017; Ogiyama et al., 2018; Schoenfelder and Fraser, 2019).
Gene regulatory networks (GRNs) specify cell identities along the dorsoventral (DV) and anterior-posterior (AP) body axes to spatially coordinate formation of the germ layers and organize the future body plan (Ma et al., 2016; Stein and Stevens, 2014). These well-characterized Drosophila GRNs are valuable for investigating how gene regulatory mechanisms converge to produce cell-type specific transcriptional programs. The importance of better understanding the molecular mechanisms of enhancer function is underscored by the presence of disease-associated variants within non-coding sequences (Rickels and Shilatifard, 2018; Zaugg et al., 2022), the pervasiveness of mutations to chromatin-modifying proteins in cancer (Flavahan et al., 2017) and the relevance of transcriptional and chromatin alterations in aging (López-Otín et al., 2013). In this review, we discuss current understanding of developmental gene regulation and particularly draw on research in Drosophila focused on uncovering the molecular mechanisms governing enhancer-promoter communication and enhancer-mediated control of transcription.
Enhancers operate in complex regulatory landscapes
Enhancer activity was initially demonstrated for a sequence element from the SV40 virus genome, capable of increasing the magnitude of transcription from the rabbit β-globin gene in HeLa cells when positioned more than 1 kb upstream or downstream from the transcription start site (TSS) (Banerji et al., 1981). Soon after, enhancer activity was detected for endogenous sequences from the eukaryotic genome (Banerji et al., 1983; Gillies et al., 1983). Enhancers have evolved to play a key role in orchestrating developmental transcriptional programs (Long et al., 2016; Shlyueva et al., 2014; Spitz and Furlong, 2012), vastly outnumbering protein-coding genes within eukaryotic genomes. The enhancer regulatory code is highly conserved across metazoans, with enhancer activities of many non-coding sequences preserved across distant species (Pennacchio et al., 2006; Wong et al., 2020). The functional hallmarks of enhancers include the capacity to modulate transcription in an orientation independent manner, operate at large genomic distances from the target promoter, and the ability to recapitulate their activity independent of the sequence context (reviewed in Shlyueva et al., 2014). In vivo reporter assay validation of sequences predicted to have enhancer activity remains essential for establishing functionality, and many candidate enhancers have not yet been validated (Kvon et al., 2014; Pennacchio et al., 2006; Shlyueva et al., 2014; Smith et al., 2023).
Chromatin restricts enhancer accessibility
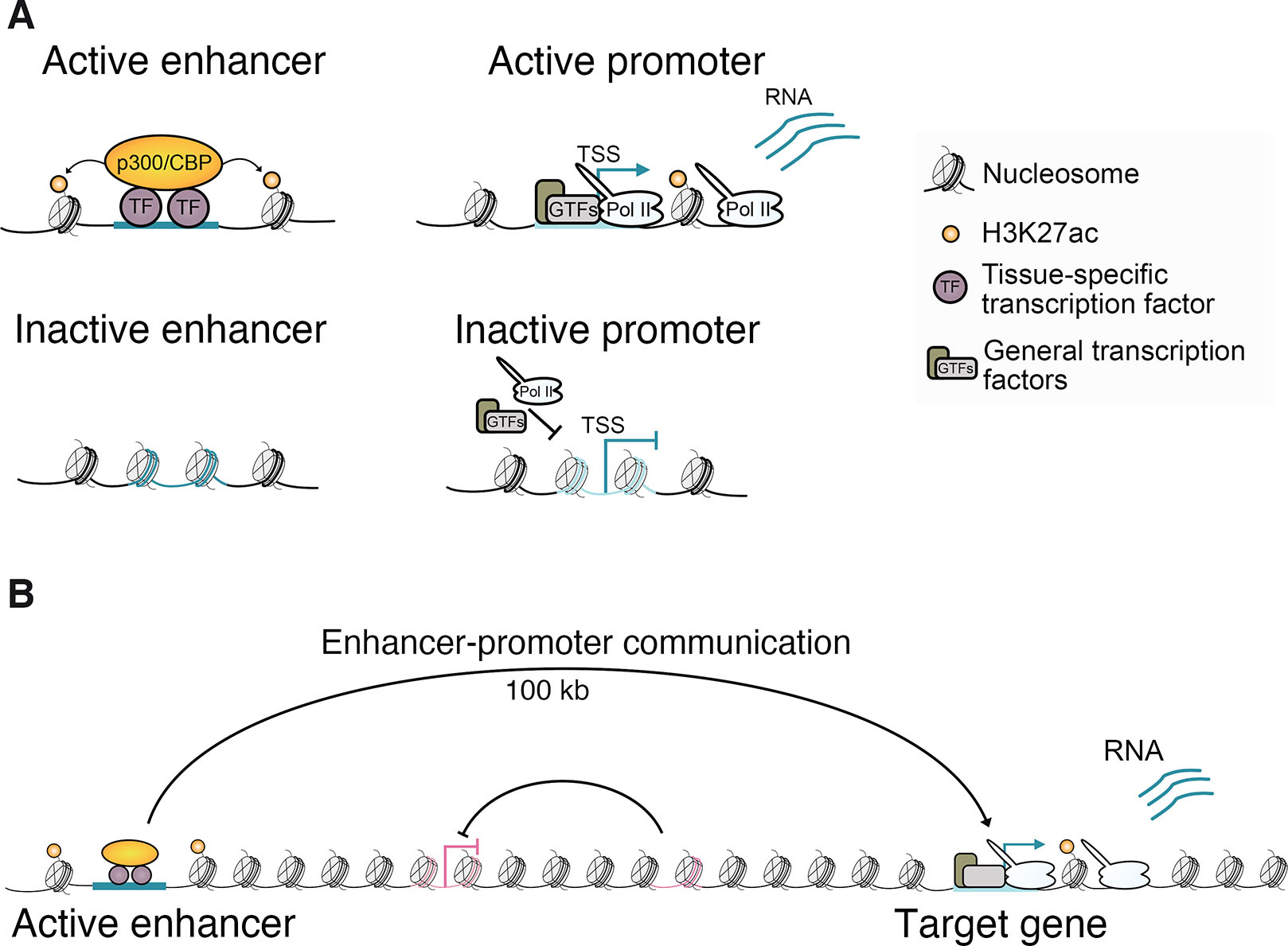
The communication of regulatory signals occurs across chromatin, and genomic approaches to predict sequences with enhancer activity based on DNA and chromatin features have been useful for identifying enhancers genome-wide and inferring cell type-specific enhancer activities (Encode Project Consortium, 2012; Negre et al., 2011; The modEncode Consortium et al., 2010). Active enhancers are characterized by accessible chromatin depleted of nucleosomes to allow for DNA binding by TFs. Genome-wide profiling has revealed that chromatin accessibility is dynamically regulated across cell types and correlates with TF binding (Fig. 2A) (Boyle et al., 2008; Li et al., 2011; Pique-Regi et al., 2011; Shlyueva et al., 2014; Spitz and Furlong, 2012; Thurman et al., 2012). Elevated chromatin accessibility is also a feature of other CREs. At active promoters, nucleosome depletion is important for assembly of the transcriptional machinery (Cairns, 2009) and Pol II may compete with nucleosomes to maintain an accessible state (Fig. 2A) (Core and Adelman, 2019; Gilchrist et al., 2010; Levine, 2011). Polycomb response elements (PREs), the recruitment sites of Polycomb group (PcG) proteins that are responsible for transcriptional silencing of developmental genes across eukaryotes (Schuettengruber et al., 2017), also exhibit low nucleosome density (Hunt et al., 2022; Mito et al., 2007; Schuettengruber et al., 2009; Schwartz et al., 2006).
Fig. 2. Enhancers operate across chromatin to influence transcription from target promoters.
(A) Schematic showing active and inactive enhancer and promoter chromatin states. (B) Communication between enhancers and promoters often occurs over large genomic distances with specificity, enabling these long-range interactions to bypass intervening enhancers and promoters.
A major function of housing the genome in chromatin is to prevent spurious transcription (Kornberg and Lorch, 2020). Therefore, factors capable of selectively producing nucleosome-depleted regions play a key role in gene regulation. A subset of TFs, known as pioneer factors, have the capacity to initiate the opening of compact chromatin (Balsalobre and Drouin, 2022; Spitz and Furlong, 2012). Pioneer factors can interact with nucleosomal DNA and recruit chromatin remodelers to deplete nucleosomes at these sites, providing DNA access for non-pioneer TFs. In this manner, the maternally supplied pioneer factor Zelda (Zld) selectively establishes chromatin accessibility at important CREs for ZGA in Drosophila (Harrison et al., 2011; Liang et al., 2008; Nien et al., 2011; Schulz and Harrison, 2019; Sun et al., 2015). Zld orchestration of ZGA is supported by several other TFs with pioneer-like activities, including the mitotic bookmarker GAGA factor (GAF) (Bellec et al., 2022; Gaskill et al., 2021), Odd-paired (Opa) (Koromila et al., 2020; Soluri et al., 2020) and CLAMP (Duan et al., 2021).
Active enhancers are marked by H3K27 acetylation
Specific signatures of histone PTMs are important markers for identifying enhancers and predicting enhancer activity states genome-wide (Shlyueva et al., 2014). Acetylation of histone H3 lysine 27 (H3K27ac) is detected at nucleosomes flanking enhancers, and its enrichment correlates with transcriptional activity from associated genes (Fig. 2A), allowing the mark to distinguish active from inactive or poised enhancers (Bonn et al., 2012; Creyghton et al., 2010; Heintzman et al., 2009; Rada-Iglesias et al., 2011). Occupancy of p300/CBP, the histone acetyltransferases responsible for depositing H3K27ac (Tie et al., 2009), has also been used to predict enhancers (Heintzman et al., 2007; Visel et al., 2009; Xi et al., 2007). Nevertheless, p300/CBP recruitment and acetyltransferase activity can be uncoupled because strong occupancy also occurs at hypoacetylated regions enriched for the repressive PcG mark tri-methylated H3K27 (H3K27me3) (Holmqvist et al., 2012; Hunt et al., 2022; Philip et al., 2015; Rada-Iglesias et al., 2011). H3K27ac marks both active enhancers and promoters, but mono- and tri-methylation of H3K4 distinguish enhancers and promoters, respectively (Heintzman et al., 2007; Rada-Iglesias et al., 2011; Shlyueva et al., 2014). Whether histone acetylation plays a direct causative role in gene activation remains largely unclear (Henikoff and Shilatifard, 2011; Millán-Zambrano et al., 2022). Histone acetylation may indirectly influence transcription by supporting the recruitment of effector proteins that recognize acetylated lysines, such as the coactivator bromodomain-containing protein 4 (BRD4) (Dey et al., 2003; Fujisawa and Filippakopoulos, 2017; Millán-Zambrano et al., 2022).
Distinct motif compositions and chromatin features shape the specificities of enhancers and promoters, influencing cofactor compatibilities (Haberle et al., 2019; Neumayr et al., 2022). Different chromatin remodelers are necessary for developmental and housekeeping transcriptional programs in Drosophila cells (Hendy et al., 2022). Remarkably, enhancers can operate long-range, spanning hundreds of kilobases (kb) from their target promoter, often bypassing intervening promoters (Fig. 2B). This implies a topological change in 3D organization, involving chromatin fiber folding, to confer specificity and allow proximity or physical contact for enhancer-promoter communication. However, while enhancers are well-defined compositionally by the TFBS they encode and specific chromatin features (Shlyueva et al., 2014; Spitz and Furlong, 2012), how signals are communicated from enhancers to target promoters, modulating transcriptional activity, remains unclear (Catarino and Stark, 2018; Furlong and Levine, 2018; Karr et al., 2022; Panigrahi and O'Malley, 2021).
Enhancer-mediated control of Drosophila developmental transcription
Studies of Drosophila embryonic patterning have unveiled how enhancers coordinate developmental gene expression (Irizarry and Stathopoulos, 2021; Small and Arnosti, 2020). The precise stripes of pair-rule gene expression across the AP axis of the early embryo are driven by multiple enhancers, each encoding different combinations of TFBS (Levine, 2010; Small and Arnosti, 2020; Small et al., 1992; Stanojevic et al., 1991). These enhancers act additively to form the overall expression pattern. The involvement of multiple enhancers with partially redundant activity, known as shadow enhancers, in directing gene expression patterns may contribute to phenotypic robustness (Cannavò et al., 2016; Frankel et al., 2010; Hong et al., 2008a; Osterwalder et al., 2018; Perry et al., 2010). Drosophila embryogenesis has been a key model for systematically characterizing non-coding sequences with developmental enhancer activity in vivo (Kvon et al., 2014). Despite its gene-dense nature relative to mammalian genomes, long-range interactions are widespread in the Drosophila genome. Many enhancer-promoter interactions occur at distances of more than 10 kb, with the majority spanning at least several kb. This separation still requires a topological mechanism to provide the necessary physical proximity for enhancer-bound TFs and cofactors to influence the transcriptional machinery at the promoter (Furlong and Levine, 2018; Ghavi-Helm et al., 2014; Hou et al., 2012; Hug et al., 2017; Sexton et al., 2012). The extensive validation of enhancers and widespread embryonic formation of enhancer-promoter interactions make Drosophila embryogenesis well suited for dissecting the mechanisms of enhancer-mediated gene activation.
Drosophila dorsal-ventral patterning
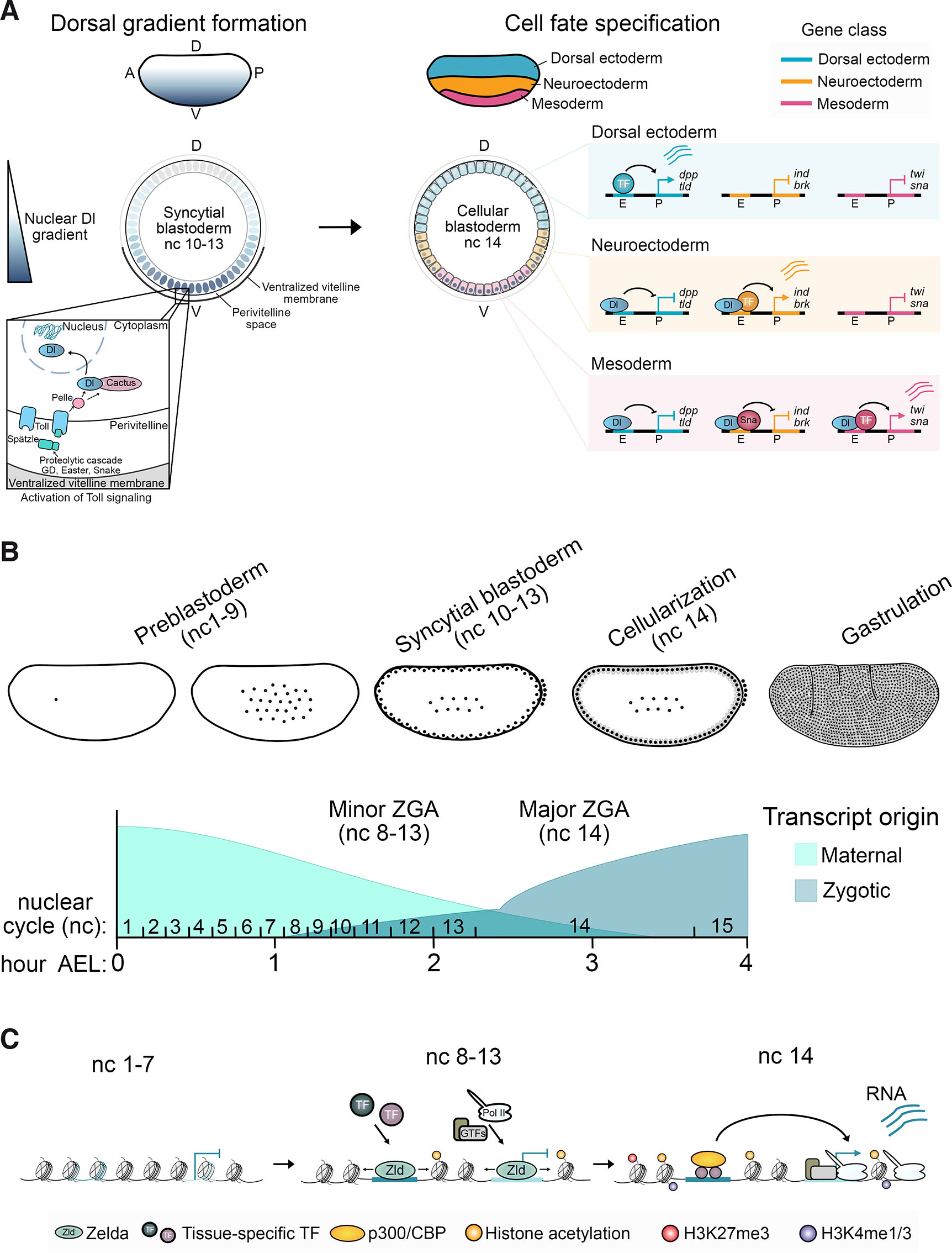
Position-dependent transcriptional programs define cell identities along the Drosophila embryonic DV axis in response to a nuclear gradient of the maternally-supplied Rel family TF Dorsal, peaking ventrally and progressively decaying dorsally (Fig. 3A) (Hong et al., 2008b; Irizarry and Stathopoulos, 2021; Stathopoulos and Levine, 2002; Stein and Stevens, 2014). A cascade of maternal effect genes relays DV polarity to the syncytial blastoderm through ventrally restricted activation of Toll signaling, releasing Dorsal from an inactive cytoplasmic complex to enter nuclei (Belvin et al., 1995; Roth et al., 1989; Rushlow et al., 1989; Steward, 1987, 1989). High nuclear Dorsal ventrally activates mesoderm-specific genes like twist (twi) and snail (sna) to form mesoderm, while intermediate and low levels of nuclear Dorsal laterally induce neuroectoderm genes, including intermediate neuroblasts defective (ind) and brinker (brk), directing neuroectoderm formation. Sna, with the CtBP and Ebi corepressors, represses neuroectoderm-specific genes in the mesoderm (Fig. 3A) (Nibu et al., 1998; Qi et al., 2008). Dorsally located cells lacking nuclear Dorsal activate genes like decapentaplegic (dpp) and tolloid (tld) for dorsal ectoderm patterning. Besides acting as a transcriptional activator for mesoderm- and neuroectoderm-specific genes, Dorsal, along with the transcriptional repressor Capicua and corepressor Groucho, confines the expression of dorsal ectoderm-specific genes to dorsally located cells (Dubnicoff et al., 1997; Papagianni et al., 2018). Dpp/BMP signaling further defines cell types along the dorsal ectoderm (Ashe et al., 2000; Ferguson and Anderson, 1992; Hamaratoglu et al., 2014). The Dorsal gradient forms during the first 90 minutes of embryogenesis and is active between nc 10-14 (Liberman et al., 2009; Reeves et al., 2012). It is during this critical developmental window that the TF cue is received by enhancers of the DV GRN and communicated to modulate transcription from the promoters of DV-regulated genes.
Fig. 3. Dorsoventral (DV) patterning of Drosophila embryogenesis and chromatin landscape establishment during zygotic genome activation (ZGA).
(A) The establishment of the Dorsal (Dl) TF nuclear gradient occurs in the syncytial blastoderm during nuclear cycle (nc) 10-13. This is facilitated by ventrally restricted activation of the Toll signaling pathway, leading to position-dependent cell fate specification of mesoderm, neuroectoderm, and dorsal ectoderm across the DV axis at nc 14. Cell identity is determined by tissue-specific transcription programs induced through enhancers responsive to different concentrations of Dl. Dorsal ectoderm-specific genes like dpp and tld are restricted to the most dorsally located cells due to Dorsal-mediated repression in lateral and ventral cells (Dubnicoff et al., 1997; Papagianni et al., 2018). Mesoderm-specific genes, including sna and twi, are expressed in ventrally-located cells with high levels of nuclear Dl, while neuroectoderm-specific genes like ind and brk are activated by Dl in lateral regions but not ventrally due to repression by Sna with the CtBP and Ebi corepressors (Nibu et al., 1998; Qi et al., 2008). Robust induction of the transcriptional programs patterning the DV axis depends on Dl acting in concert with other tissue-specific TFs, repressors, and cofactors. (B) A schematic of the stages of early Drosophila embryogenesis, illustrating nuclei (represented by black dots) dividing through rapid nuclear cycles. Nuclear divisions occur first within the center of the embryo in the preblastoderm stages (nc 1-9) before nuclei migrate to the periphery during the syncytial blastoderm stages (nc 10-13). At the periphery, nuclei cellularize to form the cellular blastoderm (nc 14) before undergoing gastrulation. The exchange of transcriptional control from maternal to zygotic during the minor and major waves of ZGA is also depicted. The hours after egg laying (AEL) are indicated across the trajectory and the corresponding nuclear cycles shown. (C) Changes to the chromatin landscape at an enhancer and promoter that become transcriptionally active at nc 14. In the lead-up to transcriptional induction, regulatory sequences are made accessible by the pioneer factor Zelda, facilitating the recruitment of TFs and the transcriptional machinery. Chromatin states that are linked to transcription, such as the H3K27ac histone modification, also begin to accumulate.
More than 100 DV-regulated genes, many encoding developmental regulators, and 200-400 enhancers have been identified. Initial interrogation of the DV GRN involved genetic analyses (reviewed in Stein and Stevens, 2014), followed by genomic approaches including bioinformatics mapping of Dorsal binding sites and whole-genome microarray analysis of the transcriptome and Dorsal occupancy in mutant embryos with uniform DV cell fates (Biemar et al., 2006; Markstein et al., 2002; Stathopoulos et al., 2002; Zeitlinger et al., 2007b). Recently, next generation sequencing methods have captured spatially- and temporally-resolved transcriptional and epigenomic landscapes during Drosophila embryogenesis (Blythe and Wieschaus, 2016; Bozek et al., 2019; Chen et al., 2013; Holmqvist et al., 2012; Hunt et al., 2024; Ing-Simmons et al., 2021; Koenecke et al., 2016; Koenecke et al., 2017; Li et al., 2014; Lott et al., 2011), including at the single-cell scale (Calderon et al., 2022; Cusanovich et al., 2018; Hunt et al., 2024; Ing-Simmons et al., 2021; Karaiskos et al., 2017; Reddington et al., 2020).
The threshold-dependent model proposes that spatially-regulated expression along the DV axis is achieved by integration of the Dorsal TF signal at enhancers responding to different Dorsal levels, determined by the affinity and organization of the binding sites they encode (Reeves and Stathopoulos, 2009; Rusch and Levine, 1996; Stathopoulos and Levine, 2002, 2004). While Dorsal initiates DV patterning, and enhancer binding site affinity correlates with positional expression (Papatsenko and Levine, 2005), precise expression domains of DV-regulated genes involve Dorsal acting in concert with other tissue-specific TFs, repressors and cofactors (Holmqvist et al., 2012; Kosman et al., 1991; Mannervik et al., 1999; Zeitlinger et al., 2007b). DV patterning constitutes one of the best-characterized GRNs in nature and has been a major model system for illuminating enhancer-mediated control of developmental transcription.
Interplay between enhancer chromatin states and tissue-specific transcription in Drosophila
During early Drosophila embryogenesis, chromatin rapidly changes in the lead-up to ZGA, transitioning from a presumed highly condensed and hypoacetylated state to a heterogeneous landscape of distinct chromatin states with selective accessibility and the deposition of histone PTMs at specific genomic regions (Fig. 3 B,C) (Blythe and Wieschaus, 2016; Bozek et al., 2019; Harrison et al., 2011; Hunt et al., 2024; Li et al., 2014; Schulz and Harrison, 2019). Histone acetylation by p300/CBP accumulates at important enhancers and promoters associated with Zld activity (Harrison et al., 2011; Hunt et al., 2024; Li et al., 2014). H3K27ac is enriched at DV enhancers and promoters, and differential H3K27ac enrichment between DV cell types has been used to predict DV-regulated enhancers (Boija and Mannervik, 2016; Ing-Simmons et al., 2021; Koenecke et al., 2016). Integrating multiple indicators of the epigenome (H3K27ac and p300/CBP enrichment and chromatin accessibility) predicted DV-regulated enhancers more accurately than each could individually (Hunt et al., 2024). Interestingly, the strength of this tissue-specific enhancer chromatin state, consisting of elevated H3K27ac, p300/CBP, and chromatin accessibility, was highly predictive of the magnitude of tissue-specific transcription from associated DV-regulated genes. The DV promoter chromatin state, as defined by these epigenomic markers, was less tissue-specific and therefore not as effective at predicting differential transcription as the enhancer. Nevertheless, while these studies have uncovered chromatin features that strongly correlate with transcriptional states, much remains ambiguous regarding functional involvement.
Functions of histone acetylation in transcription
Whether histone acetylation plays an instructive role in gene activation is unclear (Henikoff and Shilatifard, 2011; Millán-Zambrano et al., 2022). Early in vitro studies noted that histone acetylation suppressed the intrinsic inhibitory effect of histones on transcription from chromatinized DNA templates (Allfrey et al., 1964). This effect is understood to be mediated by acetylation-induced alterations to nucleosomal electrostatic interactions, leading to decompaction and increased amenability for transcription (Fenley et al., 2010). However, histone tail PTMs are considered unlikely to impact nucleosome dynamics and may instead influence transcription indirectly by providing a binding platform for effector proteins (Millán-Zambrano et al., 2022). Consistently, the coactivator BRD4 is recruited to Drosophila DV enhancers and promoters in a tissue-specific manner correlating with H3K27ac and transcriptional activation (Hunt et al., 2024). H3K27ac was also recently observed to enhance cooperative binding at human OCT4-pioneered sites (Sinha et al., 2023). Zelda binding at H3K27ac-marked DV and non-DV enhancers before ZGA in Drosophila (Hunt et al., 2024; Li et al., 2014) raises speculation about a similar association in the early embryo.
Acetylation of non-histone proteins
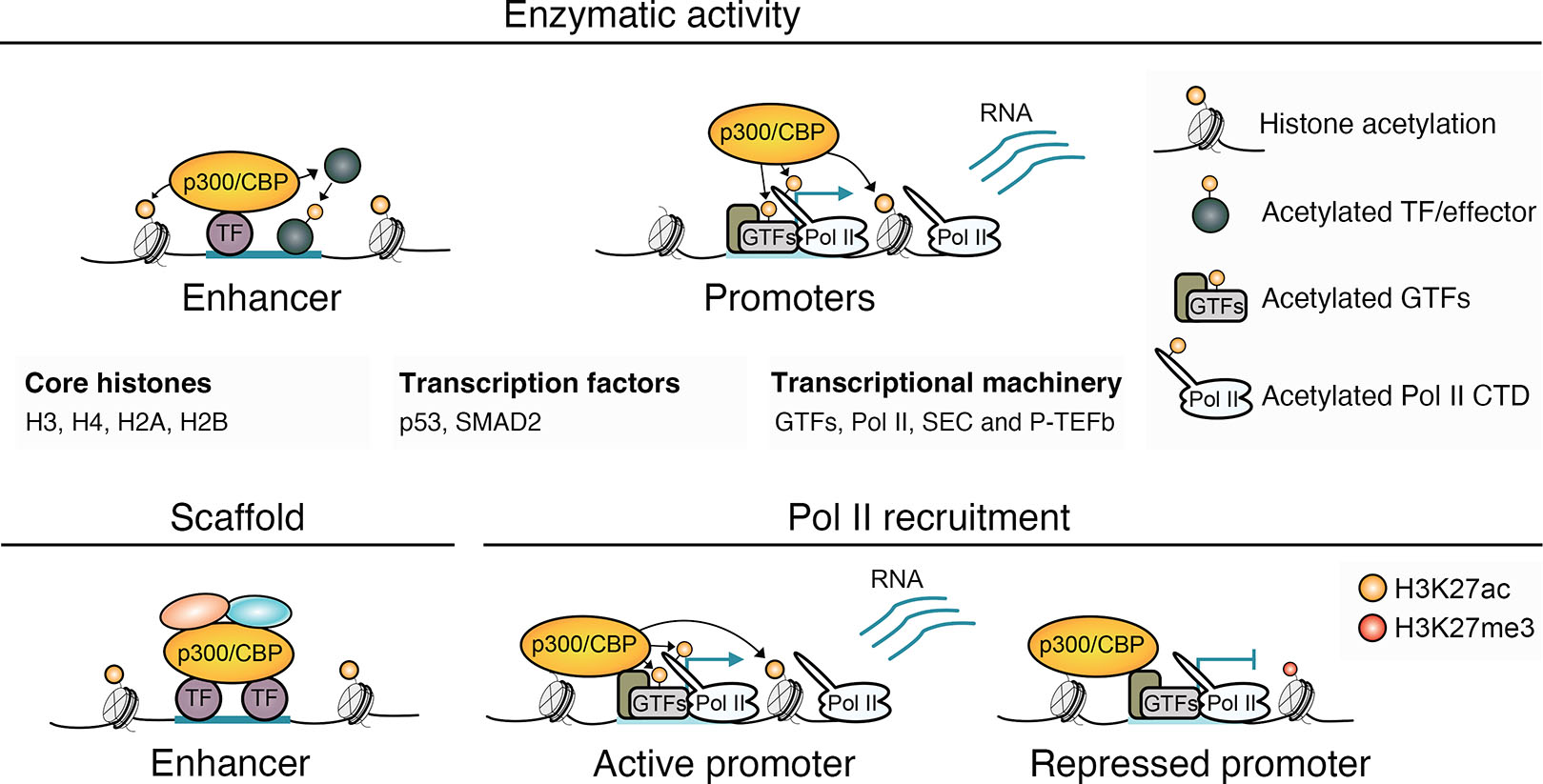
p300/CBP, like many other chromatin-modifiers, is a multifunctional protein with enzymatic and non-enzymatic activities, complicating the attribution of observed effects to specific functions (Fig. 4) (Bedford and Brindle, 2012; Dorighi et al., 2017; Hunt et al., 2022; Morgan and Shilatifard, 2020; Rickels et al., 2017). In addition to acetylating histones, the acetyltransferase activity of p300/CBP targets non-histone proteins, such as TFs, coactivators, and effectors of important signaling pathways (Weinert et al., 2018). Acetylation can influence protein function in diverse ways, including affecting protein stability, localization, protein-protein interactions, and DNA-binding ability (Spange et al., 2009). p300/CBP is equipped with a bromodomain and various other protein-protein interaction domains that may mediate non-enzymatic functions (Dancy and Cole, 2015). These functions could include acting as a scaffold to facilitate the assembly of protein complexes (Fig. 4) (Chan and La Thangue, 2001).
Fig. 4. Functions of p300/CBP in gene regulation.
The chromatin-modifier p300/CBP regulates transcription through enzymatic acetyltransferase activity, but also has non-enzymatic activities. p300/CBP acetylates lysine residues on all four core histones, including H3K27ac to mark active enhancers and promoters (Feller et al., 2015; Weinert et al., 2018), but also acetylates diverse non-histone proteins, including TFs, transcriptional regulators, and effectors of signaling pathways (Weinert et al., 2018). p300/CBP also enzymatically targets important components of the transcriptional machinery at promoters, including GTFs, Pol II, the super elongation complex (SEC) and P-TEFb, which alongside acetylation of the +1 nucleosome, may stimulate Pol II release from the promoter into productive transcription (Boija et al., 2017; Narita et al., 2021; Schröder et al., 2013; Stasevich et al., 2014). Non-enzymatic activities of p300/CBP in gene regulation could include scaffolding to help bring together TFs and other transcriptional regulators into large assemblies at target loci (Chan and La Thangue, 2001) and supporting assembly of the transcriptional machinery at promoters by interacting with TFIIB (Kwok et al., 1994). p300/CBP support of Pol II recruitment also occurs at the promoters of certain genes in repressive, H3K27me3-enriched chromatin, where productive transcription is lacking (Hunt et al., 2022).
p300/CBP is also found at promoters, where it interacts with the Pol II transcriptional machinery (Boija et al., 2017; Cho et al., 1998; Heintzman et al., 2007). At promoters, p300/CBP may influence transcription through enzymatic and non-enzymatic functions (Fig. 4) (Boija et al., 2017; Narita et al., 2021), aiding in the formation of the pre-initiation complex (PIC) via interactions with TFIIB (Kwok et al., 1994) and modulating Pol II promoter release, potentially by acetylating the transcriptional machinery and the +1 nucleosome (Boija et al., 2017; Narita et al., 2021; Schröder et al., 2013; Stasevich et al., 2014). The presence of p300/CBP and the Pol II transcriptional machinery at promoters of genes devoid of acetylation and productive transcription suggests its different functions can be partitioned (Hunt et al., 2022).
The accumulation of p300/CBP histone acetylation around ZGA is conserved in flies, zebrafish and mice (Bogdanović et al., 2012; Dahl et al., 2016; Li et al., 2014; Schulz and Harrison, 2019). Modulation of p300 acetyltransferase activity has been reported to disrupt ZGA in zebrafish (Chan et al., 2019; Miao et al., 2022; Sato et al., 2019). Interestingly, although p300/CBP is essential for ZGA in Drosophila, ZGA can occur in embryos with catalytically inactive p300/CBP (Ciabrelli et al., 2023). Moreover, p300/CBP protein depletion, but not acetyltransferase activity inhibition, compromises chromatin accessibility (Hogg et al., 2021; Hunt et al., 2022; Vannam et al., 2021), further highlighting the crucial non-enzymatic roles of p300/CBP activities in gene regulation.
Targeting H3K27 directly by replacing it with non-modifiable residues (H3K27A/R) does not compromise gene activation in Drosophila and mouse embryonic stem cells (mESCs), indicating that p300/CBP functions aside from acetylating H3K27 are important (Leatham-Jensen et al., 2019; McKay et al., 2015; Pengelly et al., 2013; Sankar et al., 2022; Zhang et al., 2020). p300/CBP is proposed to act as an "acetyl spray" that targets lysines across the tails of histones (Feller et al., 2015; Weinert et al., 2018). Supporting this spray-like activity, histone H2B tail acetylation by p300/CBP can also predict active enhancers (Narita et al., 2023). It is possible that acetylation of neighboring histone tail residues confers functional redundancy or is a by-product of enzymatic activity directed at non-histone proteins, which also affects residues on local histones. The acetyltransferase activity of p300/CBP is tightly regulated by a lysine-rich autoinhibitory loop (AIL) that, in a deacetylated state, prevents substrate access to the active site (Thompson et al., 2004). Dimerization of TFs and binding of enhancer RNAs (eRNAs) are proposed to trigger displacement of the p300/CBP AIL, allowing for enzymatic activity to occur (Bose et al., 2017; Ortega et al., 2018). The Drosophila PcG protein Pc and its mammalian CBX orthologs directly interact with the p300/CBP AIL to block acetyltransferase activity, potentially preventing turnover from transcriptionally repressive to active states (Tie et al., 2016). This aligns with the detection of p300/CBP at hypoacetylated sites (Holmqvist et al., 2012; Hunt et al., 2022; Philip et al., 2015; Rada-Iglesias et al., 2011) and supports localized multi-step enzymatic activation of p300/CBP that is uncoupled from chromatin recruitment. The transcription of DV-regulated genes anti-correlates with H3K27me3, suggesting that local enzymatic inhibition of CBP by PcGs may help ensure tissue-specific transcription (Hunt et al., 2024; Koenecke et al., 2017). CBP binding across cell types of the DV axis indicates the potential for distinct recruitment and enzymatic activity state dynamics at enhancers and promoters, where enhancer occupancy is highly tissue-specific and correlates with tissue-specific TF presence, H3K27ac, and transcription (Hunt et al., 2024). In contrast, CBP promoter binding is less tissue-specific, with residual levels persisting at promoters in inactive states.
An intriguing mechanism for enhancer-promoter communication, termed the TF activity gradient (TAG) model, proposes that acetylated TFs produced by enhancer-localized enzymatic p300/CBP activity diffuse to target promoters in close proximity, where they increase the transcriptional output (Karr et al., 2022). In this model, histone deacetylases (HDAC), chromatin modifiers that also target non-histone proteins (Glozak et al., 2005), deacetylate TFs to spatially limit the diffusing acetylated TF signal. Indeed, HDACs are recruited to active promoters in metazoans, where they may attenuate acetylated TF signals (Filion et al., 2010; Rincon-Arano et al., 2012; Wang et al., 2008). Concentrations of p300/CBP and other coactivators appear to be limiting in cells, with the number of molecules being far outnumbered by HDACs (Gillespie et al., 2020), In Drosophila cell culture, pharmacological HDAC inhibition rapidly elevates histone acetylation levels at active promoters and correlates with Pol II promoter release (Vaid et al., 2020). These findings support HDAC-mediated attenuation of acetylated TF signals as they diffuse from enhancers and the maintenance of the majority of nuclear p300/CBP in an inactive state (Karr et al., 2022).
Considering the uncertainty surrounding the direct role of histone acetylation in transcription, there is significant focus on identifying factors that undergo function-altering acetylation. This criteria may be met by the positive transcription elongation factor-b complex (P-TEFb), which stimulates Pol II elongation across gene bodies to produce transcripts (Fujinaga et al., 2023; Jonkers and Lis, 2015) and is recruited to promoters by DNA-binding TFs, including NF-κB (Barboric et al., 2001; Danko et al., 2013), and BRD4 (Dey et al., 2003; Jang et al., 2005; Yang et al., 2005). Both BRD4 and P-TEFb are recruited to Drosophila DV enhancers and promoters in a tissue-specific manner (Hunt et al., 2024) and undergo p300/CBP-dependent acetylation in mammalian cells (Weinert et al., 2018). Acetylation of P-TEFb marks its active state, free from inactive sequestration within the 7SK small nuclear ribonucleoprotein (7SK snRNP) complex (Cho et al., 2009). In summary, several observations from enhancer-driven transcription during DV patterning align with a model of enhancer-promoter communication where p300/CBP actively participates. Drosophila embryogenesis provides a conducive in vivo setting for delineating the genomic distribution of both enzymatically active and inactive p300/CBP. Further investigations can focus on unravelling the regulatory mechanisms governing the loading BRD4 and P-TEFb at promoters by enhancers, and establishing whether their acetylation occurs in a functionally significant manner.
How do enhancers influence transcriptional activity at promoters?
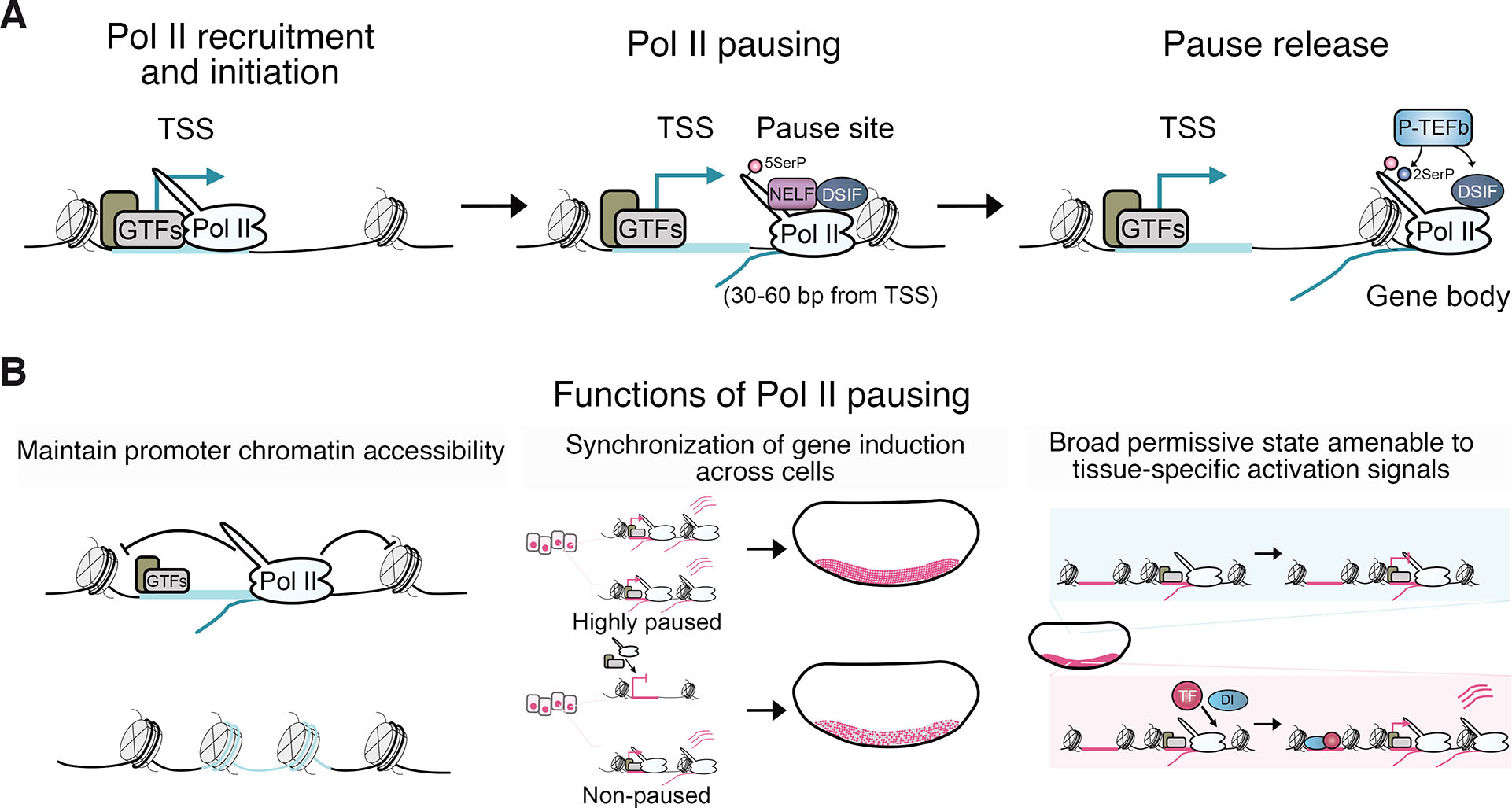
When enhancer-promoter communication occurs, signals of activation must be integrated into the transcriptional circuitry by modulating the activity of the Pol II transcriptional machinery. Transcription is a complex, multi-step process, and post-recruitment control of Pol II entry into elongation has emerged as a major rate-limiting checkpoint that controls the transcriptional output (Core and Adelman, 2019; Jonkers and Lis, 2015). At many metazoan genes, Pol II efficiently initiates transcription but pauses in the promoter-proximal region, typically 30-60 bp downstream of the TSS (Fig. 5A). This pausing of Pol II is highly stable, with most half-lives at the pause site ranging between 5-15 minutes, providing a window of time for regulatory signals from enhancers to exert their influence on the transcriptional cycle (Henriques et al., 2013; Jonkers et al., 2014; Shao and Zeitlinger, 2017). The release of Pol II from the pause site into productive elongation is tightly controlled by factors that either maintain pausing or stimulate promoter escape (Core and Adelman, 2019). P-TEFb is understood to promote pause release by phosphorylating specific residues on the Pol II C-terminal domain (CTD) and counteracting factors involved in maintaining pausing, such as Spt5, although the precise mechanism leading to elongation is not yet fully understood (Fujinaga et al., 2023; Price, 2000; Yamada et al., 2006).
Fig. 5. Pol II pausing and its functions during Drosophila embryogenesis.
(A) Schematic depicting the steps in the transcriptional cycle, showing the recruitment of Pol II, promoter-proximal Pol II pausing, and release of Pol II into productive elongation. (B) Proposed functions of Pol II pausing in gene regulation during development include aiding in repelling nucleosomes from promoter DNA to maintain accessible promoter chromatin (Gilchrist et al., 2010). Pol II pausing is also suggested to synchronize gene induction across cells of a tissue (Boettiger and Levine, 2009; Day et al., 2016; Gaertner and Zeitlinger, 2014; Lagha et al., 2013; Levine, 2011; Ramalingam et al., 2021; Saunders et al., 2013). Additionally, pausing may provide a broadly permissive promoter state that is receptive to tissue-specific signals triggering pause release and gene induction (Gaertner and Zeitlinger, 2014).
Promoter-proximal Pol II pausing at developmental genes
Drosophila early embryogenesis has played a pivotal role in elucidating the role of pausing in the coordination of tissue-specific transcription during development. Pausing was initially proposed as a mechanism to prime inducible genes for rapid signal-responsive transcriptional induction (Gilmour and Lis, 1986; Levine, 2011; Rougvie and Lis, 1988), but genome-wide mapping of Pol II occupancy subsequently uncovered the prevalence of pausing at developmental genes across metazoans (Core et al., 2008; Muse et al., 2007; Nechaev et al., 2010; Zeitlinger et al., 2007a). Consistent with serving a more fundamental and widespread role, pausing been shown to repel nucleosomes at promoters to maintain nucleosome depletion and ensure promoter DNA is kept accessible for efficient cycles of Pol II recruitment and transcription (Fig. 5B) (Core and Adelman, 2019; Gaertner and Zeitlinger, 2014; Gilchrist et al., 2010; Levine, 2011). Pausing may also help synchronize the induction of transcription across tissues and reduce transcriptional noise by attenuating variation in Pol II recruitment between cells (Boettiger and Levine, 2009; Day et al., 2016; Gaertner and Zeitlinger, 2014; Lagha et al., 2013; Levine, 2011; Ramalingam et al., 2021; Saunders et al., 2013). Highly stable paused Pol II at the pause site can inhibit new initiation events from taking place, potentially controlling the refractory period between rounds of transcription (Shao and Zeitlinger, 2017).
Pausing has been proposed to constitute a broadly permissive state across cell types that is amenable to restricted signals of gene induction (Fig. 5B) (Gaertner and Zeitlinger, 2014). While the majority of paused genes are transcriptionally active, Pol II is recruited and pauses at developmental promoters in inactive states, including DV-regulated genes during early embryogenesis (Hunt et al., 2024; Zeitlinger et al., 2007a). Pausing also occurs at the promoters of many H3K27me3-enriched PcG repressed developmental genes in mammalian embryonic stem cells (Bernstein et al., 2006; Kanhere et al., 2010; Stock et al., 2007) and Drosophila (Enderle et al., 2011; Hunt et al., 2022). Therefore, pausing appears to be a major control point of the transcriptional cycle where activating and repressing signals converge. Paused Pol II can persist at genes that have been downregulated, marking their prior activation (Gaertner et al., 2012). The presence of paused Pol II at DV-regulated genes in both active and inactive tissues seems to result from its recruitment before the establishment of the Dorsal gradient in largely transcriptionally quiescent naïve cells. These promoters may be pioneered with accessibility at this early stage of development by Zelda and are highly enriched for core promoter elements capable of recruiting the transcriptional machinery (Blythe and Wieschaus, 2016; Harrison et al., 2011; Hunt et al., 2024). These findings support tissue-specific transcription in the early embryo involving the establishment of a paused state broadly, rendering cells receptive to spatially restricted developmental cues. In the case of DV patterning, the developmental cue is tissue-specific enhancer activation by Dorsal, which may modulate BRD4 and P-TEFb activity through p300/CBP-catalyzed acetylation to promote pause release. Drosophila AP patterning genes also recruit paused Pol II in naïve cells, and mammalian Pol II is pre-loaded at promoters before ZGA (Liu et al., 2020), suggesting a conserved process for developmental gene induction.
Recently, the role of the Integrator complex in the regulation of Pol II pause release to control productive transcriptional elongation has been characterized in mammals (Fianu et al., 2021; Stein et al., 2022; Vervoort et al., 2021; Wang et al., 2023). Integrator drives premature termination through two enzymatic activities: an endonuclease that cleaves nascent RNA and a protein phosphatase that removes stimulatory phosphorylation associated with Pol II pause release and productive elongation (reviewed in Wagner et al., 2023). Targeted and rapid protein depletion has revealed that promoter H3K4me3 recruits the integrator complex subunit 11 (INTS11) endonuclease (Wang et al., 2023). Analyzing H3K4 methylation in Drosophila embryos indicates that DV enhancers acquire H3K4me1 at ZGA in both active (with H3K27ac) and uninduced (without H3K27ac) states, while both H3K4me1 and H3K4me3 are present at DV promoters (Koenecke et al., 2017; Li et al., 2014). It would be interesting to apply the DV model system to distinguish cell type-specific differences in the deposition of H3K4 methylation and functionally interrogate the relationship between tissue-specific chromatin features and pause release during development.
Transcription occurs in bursts that are modulated during developmental gene regulation
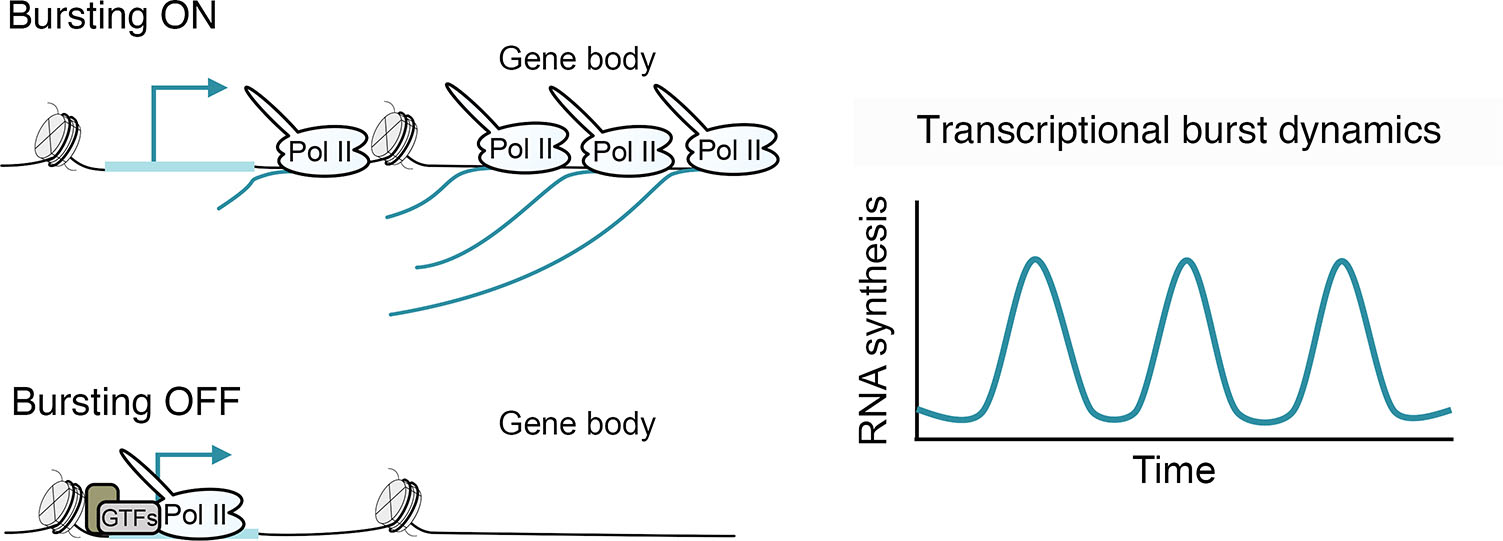
Transcription occurs in stochastic bursts of RNA synthesis, resulting from alternation between active and inactive states (Fig. 6) (Leyes Porello et al., 2023; Rodriguez and Larson, 2020; Tunnacliffe and Chubb, 2020). The discontinuous nature of transcription was first observed in Drosophila embryo chromosome spreads by electron microscopy in the late 1970s (Miller Jr. and McKnight, 1979). Since then, the ability to study gene bursting has been spurred by the development of highly sensitive microscopy and genomics techniques that have been leveraged to explore transcriptional dynamics at the single-cell scale. Drosophila provides a major model system for studying gene bursting in vivo during development. Bursting has been visualized for genes of interest in early Drosophila embryos by single-molecule RNA FISH (smFISH) and captured live in real-time using the MS2/MCP system (Leyes Porello et al., 2023). Gene bursting dynamics have also been inferred transcriptome-wide from single-cell RNA-seq (scRNA-seq) data (Hunt et al., 2024; Jiang et al., 2017; Kim and Marioni, 2013; Larsson et al., 2019).
Several models, differing in the number of promoter activity states accommodated, have been applied to describe transcriptional bursts (Rodriguez and Larson, 2020; Tunnacliffe and Chubb, 2020). A two-state model, where promoters fluctuate between active/ON and inactive/OFF configurations, has been widely adopted to infer transcriptional dynamics (Berrocal et al., 2020; Larsson et al., 2019; Peccoud and Ycart, 1995; Raj et al., 2006; Zoller et al., 2018). Bursts have been characterized by parameters including frequency and size (number of RNA molecules produced per burst) that can be inferred from this model. However, the failure to accommodate intermediary promoter activity states, such as pausing, means the two-state model may not always satisfactorily capture promoter dynamics (Tunnacliffe and Chubb, 2020). As a result, multistate models have been proposed to better explain the transcriptional activity of some genes (Bartman et al., 2019; Corrigan et al., 2016; Neuert et al., 2013; Pimmett et al., 2021; Suter et al., 2011). While these experimental techniques and modelling approaches have helped establish that gene burstiness is a highly evolutionarily conserved feature of transcription, a major focus has been to reveal the molecular determinants that modulate bursting dynamics.
Studies of the molecular determinants of bursts have linked enhancers to transcriptional control through modulation of gene burst frequencies (Fukaya et al., 2016; Walters et al., 1995). Chromatin features, such as differential histone acetylation at enhancers, have been found to correlate with the burst frequency of target genes (Larsson et al., 2019; Nicolas et al., 2018). The burst size of genes shows a strong association with the occurrence of specific core promoter motifs, suggesting that it is largely sequence-encoded at promoters (Hornung et al., 2012; Larsson et al., 2019). Inferring burst kinetics in Drosophila embryonic DV tissue scRNA-seq data corroborates that enhancer-mediated control of burst frequencies and promoter-encoded burst sizes underlie developmental gene induction in vivo (Hunt et al., 2024). DV genes are characterized by a low burst frequency and the capacity for strong bursts. The high burst size capacity may derive from an overrepresentation of the core promoter motifs TATA and INR that bind TFIID to trigger PIC assembly (Haberle and Stark, 2018; Joo et al., 2017), but could possibly also be influenced by pausing (Hunt et al., 2024; Pimmett et al., 2021; Tantale et al., 2021). Notably, differential promoter loading of P-TEFb between DV tissues correlates with burst size, implicating the pause release circuitry in the induction of strong bursts (Hunt et al., 2024). The tissue-specific active chromatin state found at DV enhancers correlates strongly with elevated burst frequencies at target promoters but is also well correlated with burst size increases for genes whose induction does not involve significant changes in burst frequency. While the molecular determinants that modulate bursts are being revealed, much remains unclear about the mechanism of enhancer-promoter communication that allows burst induction to be signaled.
Enhancer-promoter communication across the chromatin landscape
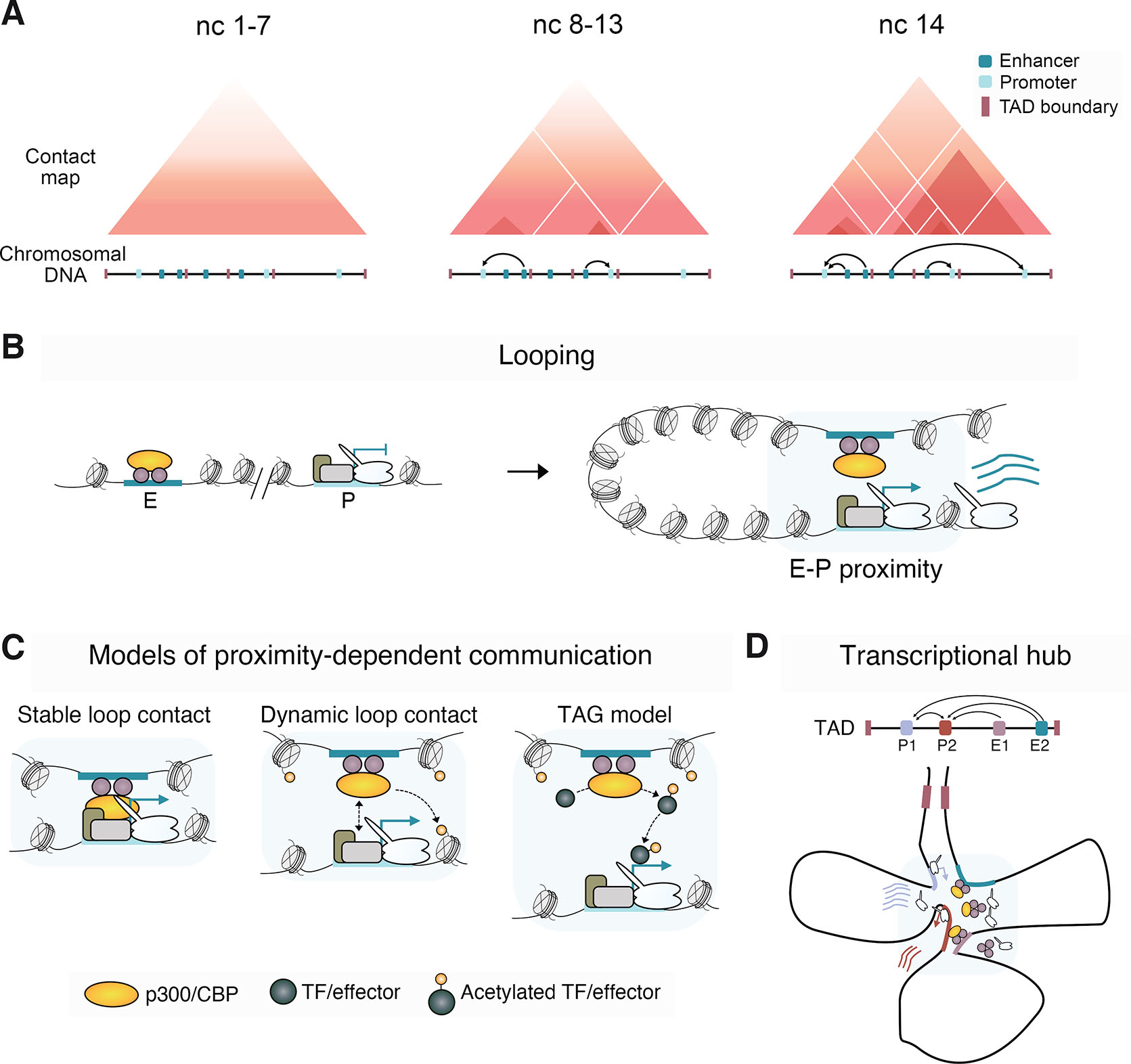
Enhancer-promoter communications occur in complex regulatory environments where they must overcome the challenges of genomic distance and specificity when surrounded by non-target enhancers and promoters. These communications occur within the genomic organization of TADs (Dixon et al., 2012; Sexton et al., 2012), with boundaries harboring insulator activities that prevent unintended interactions between enhancers and promoters in different TADs (Jerkovic and Cavalli, 2021). Similar to their mammalian counterparts, Drosophila TAD boundaries are enriched in DNA binding motifs for insulator proteins (Hou et al., 2012; Hug et al., 2017; Jiang et al., 2009; Kaushal et al., 2022; Sexton et al., 2012; Stadler et al., 2017; Ulianov et al., 2016). Techniques that capture chromosome conformation have revealed the emergence of Drosophila TADs and focal contacts between enhancers and promoters in parallel with ZGA (Fig. 7A) (Ghavi-Helm et al., 2014; Hug et al., 2017; Ogiyama et al., 2018). In addition to containing motifs for insulator proteins, early forming Drosophila TAD boundaries have specific signatures of histone modifications and can be enriched for Pol II and Zld (Hug et al., 2017). TADs form independently of zygotic transcription, but insulation at some boundaries is compromised by transcriptional inhibition and depletion of Zld.
Fig. 7. 3D genome organization and models of enhancer-promoter communication.
(A) Schematic of genome organization during early Drosophila embryogenesis, showing the formation of TADs and the establishment of enhancer-promoter contacts. (B) Model presenting the looping topological conformation that brings enhancers and promoters into proximity. (C) Models illustrating proximity-dependent enhancer-promoter communication, showing stable and transient direct enhancer-promoter contacts mediated by physical interactions between enhancer- and promoter-bound factors. Dynamic interactions may transmit signals to promoters via the deposition of histone PTMs at promoter chromatin by enhancer-bound chromatin factors. The transcription factor activity gradient (TAG) model suggests an enhancer-localized activation signal, such as p300/CBP-mediated acetylation of TFs or transcriptional effector like P-TEFb, diffuses to the target promoter in close proximity to induce transcription (Karr et al., 2022). (D) The transcriptional hub model for gene activation proposes the clustering of multiple enhancers and promoters in proximity. Within this hub environment, a pool of high concentrations of TFs, cofactors, and Pol II collectively drives robust activation (Furlong and Levine, 2018).
In mammals and Drosophila, there is disparity between the loss of TAD insulation when insulator proteins are perturbed and the modest effects on ensuing gene expression (Cavalheiro et al., 2023; Kaushal et al., 2021; Nora et al., 2017; Rao et al., 2017; Schwarzer et al., 2017). Disruption of TAD boundaries in some instances have been demonstrated to rewire genome organization, leading to ectopic enhancer-promoter interactions that impact gene expression in mammals (Franke et al., 2016; Lupiáñez et al., 2015; Nora et al., 2012). However, the depletion of architectural proteins understood to mediate contacts and the dramatically rearranged genome organization in Drosophila balancer chromosomes do not majorly affect enhancer-promoter communications and transcription (Ghavi-Helm et al., 2019; Hsieh et al., 2022). While TADs are reported to be largely invariant between cell types (Dixon et al., 2015; Dixon et al., 2012; Nora et al., 2012), cell type-specific TADs have been detected, and intra-TAD interactions are reported to be dynamically regulated (Bonev et al., 2017; Chathoth and Zabet, 2019; Kragesteen et al., 2018; Le Dily et al., 2014; Mateo et al., 2019). A class of CREs known as tethering elements has been shown to facilitate intra-TAD enhancer-promoter and promoter-promoter interactions in Drosophila (Batut et al., 2022; Calhoun et al., 2002). Moreover, GAF-mediated pairing of tethering elements may help ensure CRE selectivity (Batut et al., 2022; Li et al., 2023). Promoter-promoter contacts between promoter-proximal tethering elements have been shown to underlie the transcriptional coupling of genomically distant functionally-related genes with similar spatio-temporal expression dynamics driven by shared enhancers (Levo et al., 2022). Some interactions between enhancers and promoters also occur across TADs (Batut et al., 2022; Balasubramanian et al., 2024; Yokoshi et al., 2020).
While TADs are proposed to produce insulated genomic domains within which enhancers and promoters interact, the mechanisms behind these interactions remain unclear (Furlong and Levine, 2018). Chromatin looping is proposed to bring genomically distant enhancers and promoters into contact (Fig. 7B). The formation of chromatin loops may involve physical interactions between enhancer and promoter-bound architectural proteins, such as CTCF, and TFs with stable or transient proximity required for transcriptional activation (Fig. 7C). Both population-based chromosome conformation methods and imaging have observed enhancers and promoters in close physical proximity (Cruz-Molina et al., 2017; Espinola et al., 2021; Ghavi-Helm et al., 2014; Hsieh et al., 2020; Ing-Simmons et al., 2021; Jin et al., 2013; Krietenstein et al., 2020; Li et al., 2012; Sanyal et al., 2012). Generally, enhancer-promoter proximity correlates with gene activity (Bonev et al., 2017; Ghavi-Helm et al., 2014; Mateo et al., 2019; Rao et al., 2014; Sanyal et al., 2012), and forced looping can induce transcription of some genes (Bartman et al., 2016; Deng et al., 2012; Deng et al., 2014). However, certain studies have detected no correlation, or even an anti-correlation, between enhancer-promoter proximity and gene activity (Alexander et al., 2019; Benabdallah et al., 2019; Chen et al., 2018; Heist et al., 2019). The model of dedicated and stable looping between an enhancer and its target promoter is challenged by the capacity of an enhancer to simultaneously drive bursts of transcription from flanking genes and to co-activate reporter genes in cis and in trans through the phenomenon of transvection in Drosophila embryos (Fukaya et al., 2016; Lim et al., 2018).
An alternative model of enhancer-promoter communication, not reliant on direct looping, is the TF activity gradient (TAG), proposing that acetylated TFs produced by enhancer-localized enzymatic p300/CBP activity diffuse to target promoters in close proximity to activate transcription (Fig. 7C) (Karr et al., 2022). In this model, histone deacetylases (HDACs), chromatin modifiers that also enzymatically target non-histone proteins (Glozak et al., 2005), deacetylate TFs to spatially limit the diffusing acetylated TF signal. HDACs are recruited to active promoters in metazoans, where they may attenuate acetylated TF signals (Filion et al., 2010; Rincon-Arano et al., 2012; Wang et al., 2008). The quantity of molecules from coactivators such as p300/CBP appears to be limited in cells, being far surpassed by HDACs (Gillespie et al., 2020). Consistently, pharmacological HDAC inhibition in Drosophila cells rapidly elevates histone acetylation levels at active promoters and correlates with Pol II promoter release (Vaid et al., 2020). Overall, the relationship between enhancer-promoter proximity and the transfer of regulatory cues that lead to activation remains unresolved.
Gene activation in invariant chromosome conformations
During Drosophila embryogenesis, enhancer-promoter proximities are established before transcriptional activation (Espinola et al., 2021; Ghavi-Helm et al., 2014). Strikingly, despite substantial differences in chromatin state and transcriptional activity, DV loci do not markedly change in chromatin conformation between DV tissues (Ing-Simmons et al., 2021). High-resolution imaging of CRE interactions within the dorsocross (doc) TAD, which encompasses three co-expressed dorsal ectoderm-specific genes, revealed that multiple enhancers and promoters coalesce in close proximity independently of cell fate (Espinola et al., 2021). Similarly, anterior-posterior regulated genes display a consistent chromatin conformation in both anterior and posterior halves of early Drosophila embryos, despite their distinct transcriptional states (Stadler et al., 2017). These observations suggest that a preformed topology with close proximity between enhancers and promoters might be a crucial prerequisite for gene activity, but additional signals are necessary to initiate transcription. Interestingly, promoters associated with preformed interactions are enriched for paused Pol II, further implying their priming for activity in response to the required regulatory cue (Ghavi-Helm et al., 2014).
Nuclear hubs and phase separation in gene activation
Observations of close proximity between potentially multiple enhancers and promoters are consistent with the concept of nuclear hubs or transcription factories, where high concentrations of TFs, cofactors, and the Pol II machinery drive robust bursts of gene expression (Fig. 7D) (Furlong and Levine, 2018; Iborra et al., 1996; Lim and Levine, 2021). Zelda forms sub-nuclear hubs of high concentration within which frequent transient binding events occur that potentiate gene expression during Drosophila embryonic patterning (Dufourt et al., 2018; Mir et al., 2018). Notably, the size of enhancer TF clusters of the anterior-posterior patterning morphogen Bicoid correlates with the burst size of target reporter genes (Kawasaki and Fukaya, 2023).
The process of liquid-liquid phase separation (LLPS), where dense cooperative assemblies of proteins form liquid-like condensates or droplets, has been proposed to explain how high sub-nuclear concentrations of transcriptional components might be achieved (Hnisz et al., 2017). Condensate formation is thought to be driven by multivalent interactions through intrinsically disordered regions (IDRs). Liquid-like condensate formation has been associated with the transcriptional capacity of several coactivators (Boija et al., 2018; Cho et al., 2018; Sabari et al., 2018) and Pol II (Boehning et al., 2018; Cho et al., 2018; Guo et al., 2019; Lu et al., 2018), which have consistently been detected in nuclear clusters by various methods (Cisse et al., 2013; Cook, 1999; Pownall et al., 2023). p300/CBP nuclear condensates are linked to enhanced HAT activity at active genomic sites (Ma et al., 2021) but also the sequestration of HAT inactive condensates in H3K27me3-enriched chromatin (Zhang et al., 2021).
Condensates are also influenced by DNA sequence, with enhancer elements dense in TF recruitment motifs supporting condensate formation in vitro (Shrinivas et al., 2019). While IDR interactions may underlie TF and Pol II clustering (Chong et al., 2018), much debate concerns whether cluster formation in vivo is attributed to LLPS and what relevance this may pertain to transcription (Alberti et al., 2019; McSwiggen et al., 2019). Multivalent clustering has been shown to enhance the transcription activation strength of synthetic TFs, but liquid-like droplet formation did not (Trojanowski et al., 2022).
The formation of localized clusters of TFs, cofactors and the Pol II machinery into hubs at enhancers and promoters in close proximity presents a plausible organization for gene regulatory circuitry. Interestingly, RNAs associated with transcription initiation stimulate condensate formation, while RNAs produced during bursts of elongation have been linked to condensate dissociation (Henninger et al., 2021). This observation aligns with Pol II cluster dynamics observed through chromatin expansion microscopy, where enhancer-promoter interactions occur transiently, and bursts of transcriptional elongation are associated with their separation, termed the "kiss and kick" model (Pownall et al., 2023). At present, what constitutes the transmissible enhancer signal is unclear.
Conclusion
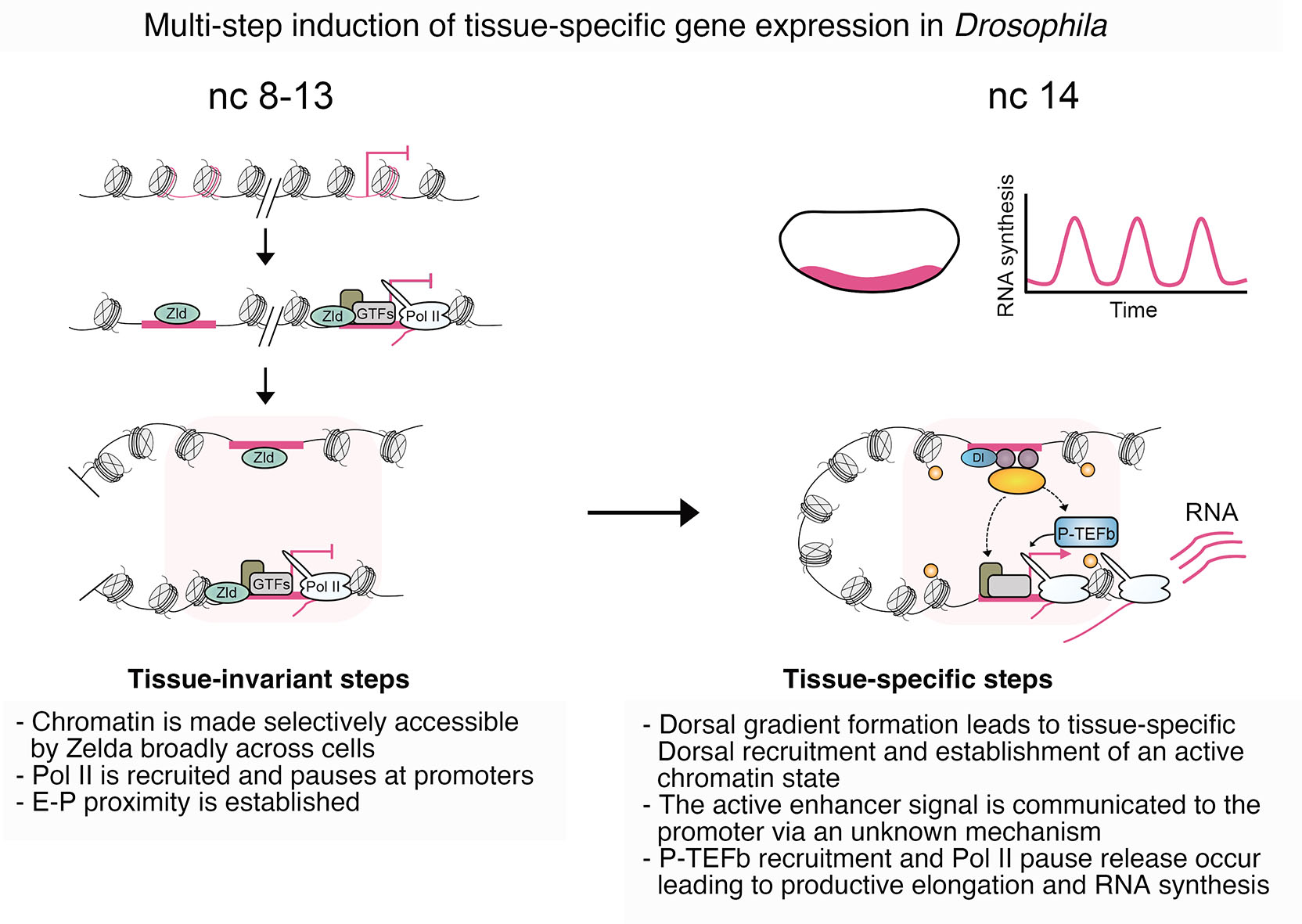
Drosophila embryogenesis has served as a key model system for functionally dissecting developmental gene regulation. The rapid induction of cell type-specific transcriptional programs at ZGA well positions the Drosophila embryo for studying the molecular mechanisms governing how enhancers communicate regulatory cues to promoters and induce transcription. Leading up to ZGA, an elaborate genome organization forms with differential chromatin accessibility, TF binding, and histone PTM deposition, along with the establishment of proximity between enhancers and promoters. The interaction of these different layers of genome regulation to direct transcription across space and time remains unresolved. A major goal is to obtain a mechanistic understanding of how regulatory information is transferred between enhancers and promoters and what the communicable signal may be. It is striking that chromosome conformation is similar in different DV cell types, despite their major differences in chromatin and transcriptional states. This suggests a preformed topology may be a prerequisite, but not the molecular trigger, of enhancer-promoter communication. Such an organization could facilitate multi-step regulation of developmental transcription, as seen in the DV GRN. Here, within a pre-established organizational architecture of enhancer-promoter proximity, TFs and cofactors may cluster at enhancers in a cell type-specific manner to initiate bursts of transcription from promoters primed with paused Pol II (Fig. 8). The DV GRN is well-positioned for future efforts to functionally interrogate potential mediators of communication, such as rapid optogenetic depletion of p300/CBP or effectors it may modify, like BRD4 or P-TEFb, as achieved in the early embryo for other factors (Huang et al., 2017; McDaniel et al., 2019; Singh et al., 2022).
Fig. 8. Developmental gene induction during Drosophila developmental patterning.
The current understanding of Drosophila DV gene regulation suggests a multi-step process in the activation of DV-regulated genes. Chromatin is made accessible by Zelda at key enhancers and promoters. Following this, Pol II pausing is established, and enhancer-promoter proximities form across cells prior to ZGA (Hunt et al., 2024). Tissue-specific signals, in the form of the Dorsal (Dl) TF gradient and other tissue-specific TFs, lead to the formation of an active chromatin state. The communication of this signal to target promoters occurs via an unknown mechanism, ultimately inducing the release of paused Pol II from target promoters.
A key aspect of elucidating enhancer-promoter communication is pinpointing the steps in the transcriptional cycle where regulatory influence is integrated. The DV GRN suggests that tissue-specific transcription is driven by post-recruitment control of the transition of Pol II from pausing to elongating. The recruitment of paused Pol II across cells in the early embryo suggests the widespread adoption of a promoter state conducive to tissue-specific signals. Enhancer-driven transcriptional induction, achieved through control of pause release, is consistent with modulation of the pause release machinery being a key output of enhancer activation. Drosophila embryogenesis is primed to continue to play a pivotal role in unravelling these unknowns.
Acknowledgements
We thank members of the Mannervik lab for helpful discussions. The work was funded through the Swedish Research Council (Vetenskapsrådet) to M.M.
Abbreviations
AIL, Autoinhibitory loop ; AP, Anterior-posterior ; CREs, Cis-regulatory elements ; Dl, Dorsal ; DV, Dorsoventral ; GRNs, Gene regulatory networks ; GTFs, General transcription factors ; H3K27ac, Histone H3 lysine 27 acetylation ; H3K27me3, Histone H3 lysine 27 tri-methylation ; nc, nuclear cycle ; Pol II, RNA polymerase II ; PTMs, Posttranslational modifications ; TADs, Topologically associating domains ; TF, Transcription factor ; TFBS, TF binding sites ; TSS, Transcription start site ; ZGA, Zygotic genome activation ;Declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
Alberti S., Gladfelter A., Mittag T. (2019). Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176: 419-434.
Alexander J. M., Guan J., Li B., Maliskova L., Song M., Shen Y., Huang B., Lomvardas S., Weiner O. D., (2019). Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife 8: e41769.
Allfrey V. G., Faulkner R., Mirsky A. E. (1964). ACETYLATION AND METHYLATION OF HISTONES AND THEIR POSSIBLE ROLE IN THE REGULATION OF RNA SYNTHESIS. Proceedings of the National Academy of Sciences 51: 786-794.
Ashe H. L., Mannervik M., Levine M. (2000). Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development 127: 3305-3312.
Balasubramanian D., Borges Pinto P., Grasso A., Vincent S., Tarayre H., Lajoignie D., Ghavi-Helm Y. (2024). Enhancer–promoter interactions can form independently of genomic distance and be functional across TAD boundaries. Nucleic Acids Research 52: 1702-1719.
Balsalobre A., Drouin J. (2022). Pioneer factors as master regulators of the epigenome and cell fate. Nature Reviews Molecular Cell Biology 23: 449-464.
Banerji J., Olson L., Schaffner W. (1983). A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell 33: 729-740.
Banerji J., Rusconi S., Schaffner W. (1981). Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell 27: 299-308.
Bannister A. J., Kouzarides T. (2011). Regulation of chromatin by histone modifications. Cell Research 21: 381-395.
Barboric M., Nissen R. M., Kanazawa S., Jabrane-Ferrat N., Peterlin B.M. (2001). NF-κB Binds P-TEFb to Stimulate Transcriptional Elongation by RNA Polymerase II. Molecular Cell 8: 327-337.
Bartman C. R., Hamagami N., Keller C. A., Giardine B., Hardison R. C., Blobel G. A., Raj A. (2019). Transcriptional Burst Initiation and Polymerase Pause Release Are Key Control Points of Transcriptional Regulation. Molecular Cell 73: 519-532.e4.
Bartman C. R., Hsu S. C., Hsiung C. C.S., Raj A., Blobel G. A. (2016). Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Molecular Cell 62: 237-247.
Batut P. J., Bing X. Y., Sisco Z., Raimundo J., Levo M., Levine M. S. (2022). Genome organization controls transcriptional dynamics during development. Science 375: 566-570.
Bedford D. C., Brindle P. K. (2012). Is histone acetylation the most important physiological function for CBP and p300?. Aging 4: 247-255.
Bellec M., Dufourt J., Hunt G., Lenden-Hasse H., Trullo A., Zine El Aabidine A., Lamarque M., Gaskill M. M., Faure-Gautron H., Mannervik M., Harrison M. M., Andrau J.C., Favard C., Radulescu O., Lagha M. (2022). The control of transcriptional memory by stable mitotic bookmarking. Nature Communications 13: 1176.
Belvin M. P., Jin Y., Anderson K. V. (1995). Cactus protein degradation mediates Drosophila dorsal-ventral signaling.. Genes & Development 9: 783-793.
Benabdallah N. S., Williamson I., Illingworth R. S., Kane L., Boyle S., Sengupta D., Grimes G. R., Therizols P., Bickmore W. A. (2019). Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Molecular Cell 76: 473-484.e7.
Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006). A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 125: 315-326.
Berrocal A., Lammers N. C., Garcia H. G., Eisen M. B. (2020). Kinetic sculpting of the seven stripes of the Drosophila even-skipped gene. eLife 9: e61635.
Biemar F., Nix D. A., Piel J., Peterson B., Ronshaugen M., Sementchenko V., Bell I., Manak J. R., Levine M. S. (2006). Comprehensive identification of Drosophila dorsal–ventral patterning genes using a whole-genome tiling array. Proceedings of the National Academy of Sciences 103: 12763-12768.
Blythe S. A., Wieschaus E. F. (2016). Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife 5: e20148.
Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A. S., Yu T., Marie-Nelly H., McSwiggen D. T., Kokic G., Dailey G. M., Cramer P., Darzacq X., Zweckstetter M. (2018). RNA polymerase II clustering through carboxy-terminal domain phase separation. Nature Structural & Molecular Biology 25: 833-840.
Boettiger A. N., Levine M. (2009). Synchronous and Stochastic Patterns of Gene Activation in the Drosophila Embryo. Science 325: 471-473.
Bogdanović O., Fernandez-Miñán A., Tena J. J., de la Calle-Mustienes E., Hidalgo C., van Kruysbergen I., van Heeringen S. J., Veenstra G. J. C., Gómez-Skarmeta J. L. (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Research 22: 2043-2053.
Boija A., Klein I. A., Sabari B. R., Dall’Agnese A., Coffey E. L., Zamudio A. V., Li C. H., Shrinivas K., Manteiga J. C., Hannett N. M., Abraham B. J., Afeyan L. K., Guo Y. E., Rimel J. K., Fant C. B., Schuijers J., Lee T. I., Taatjes D. J., Young R. A. (2018). Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175: 1842-1855.e16.
Boija A., Mahat D. B., Zare A., Holmqvist P.H., Philip P., Meyers D. J., Cole P. A., Lis J. T., Stenberg P., Mannervik M. (2017). CBP Regulates Recruitment and Release of Promoter-Proximal RNA Polymerase II. Molecular Cell 68: 491-503.e5.
Boija A., Mannervik M. (2016). Initiation of diverse epigenetic states during nuclear programming of the Drosophila body plan. Proceedings of the National Academy of Sciences 113: 8735-8740.
Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G. L., Lubling Y., Xu X., Lv X., Hugnot J.P., Tanay A., Cavalli G. (2017). Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171: 557-572.e24.
Bonn S., Zinzen R. P., Girardot C., Gustafson E. H., Perez-Gonzalez A., Delhomme N., Ghavi-Helm Y., Wilczyński B., Riddell A., Furlong E. E. M. (2012). Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nature Genetics 44: 148-156.
Bose D. A., Donahue G., Reinberg D., Shiekhattar R., Bonasio R., Berger S. L. (2017). RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 168: 135-149.e22.
Boyle A. P., Davis S., Shulha H. P., Meltzer P., Margulies E. H., Weng Z., Furey T. S., Crawford G. E. (2008). High-Resolution Mapping and Characterization of Open Chromatin across the Genome. Cell 132: 311-322.
Bozek M., Cortini R., Storti A. E., Unnerstall U., Gaul U., Gompel N. (2019). ATAC-seq reveals regional differences in enhancer accessibility during the establishment of spatial coordinates in the Drosophila blastoderm. Genome Research 29: 771-783.
Cairns B. R. (2009). The logic of chromatin architecture and remodelling at promoters. Nature 461: 193-198.
Calderon D., Blecher-Gonen R., Huang X., Secchia S., Kentro J., Daza R. M., Martin B., Dulja A., Schaub C., Trapnell C., Larschan E., O’Connor-Giles K. M., Furlong E. E. M., Shendure J. (2022). The continuum of Drosophila embryonic development at single-cell resolution. Science 377: eabn5800.
Calhoun V. C., Stathopoulos A., Levine M. (2002). Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proceedings of the National Academy of Sciences 99: 9243-9247.
Cannavò E., Khoueiry P., Garfield D. A., Geeleher P., Zichner T., Gustafson E. H., Ciglar L., Korbel J. O., Furlong E. E.M. (2016). Shadow Enhancers Are Pervasive Features of Developmental Regulatory Networks. Current Biology 26: 38-51.
Catarino R. R., Stark A. (2018). Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes & Development 32: 202-223.
Cavalheiro G. R., Girardot C., Viales R. R., Pollex T., Cao T. B. N., Lacour P., Feng S., Rabinowitz A., Furlong E. E. M. (2023). CTCF, BEAF-32, and CP190 are not required for the establishment of TADs in early Drosophila embryos but have locus-specific roles. Science Advances 9: eade1085.
Chan H. M., La Thangue N. B. (2001). p300/CBP proteins: HATs for transcriptional bridges and scaffolds. Journal of Cell Science 114: 2363-2373.
Chan S. H., Tang Y., Miao L., Darwich-Codore H., Vejnar C. E., Beaudoin J.D., Musaev D., Fernandez J. P., Benitez M. D.J., Bazzini A. A., Moreno-Mateos M. A., Giraldez A. J. (2019). Brd4 and P300 Confer Transcriptional Competency during Zygotic Genome Activation. Developmental Cell 49: 867-881.e8.
Chathoth K. T., Zabet N. R. (2019). Chromatin architecture reorganization during neuronal cell differentiation in Drosophila genome. Genome Research 29: 613-625.
Chen H., Levo M., Barinov L., Fujioka M., Jaynes J. B., Gregor T. (2018). Dynamic interplay between enhancer–promoter topology and gene activity. Nature Genetics 50: 1296-1303.
Chen K., Johnston J., Shao W., Meier S., Staber C., Zeitlinger J. (2013). A global change in RNA polymerase II pausing during the Drosophila midblastula transition. eLife 2: e00861.
Cho H., Orphanides G., Sun X., Yang X.J., Ogryzko V., Lees E., Nakatani Y., Reinberg D. (1998). A Human RNA Polymerase II Complex Containing Factors That Modify Chromatin Structure. Molecular and Cellular Biology 18: 5355-5363.
Cho S., Schroeder S., Kaehlcke K., Kwon H.S., Pedal A., Herker E., Schnoelzer M., Ott M. (2009). Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. The EMBO Journal 28: 1407-1417.
Cho W.K., Spille J.H., Hecht M., Lee C., Li C., Grube V., Cisse I. I. (2018). Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361: 412-415.
Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G. M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., Tjian R. (2018). Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361: eaar2555.
Ciabrelli F., Rabbani L., Cardamone F., Zenk F., Löser E., Schächtle M. A., Mazina M., Loubiere V., Iovino N. (2023). CBP and Gcn5 drive zygotic genome activation independently of their catalytic activity. Science Advances 9: eadf2687.
Cisse I. I., Izeddin I., Causse S. Z., Boudarene L., Senecal A., Muresan L., Dugast-Darzacq C., Hajj B., Dahan M., Darzacq X. (2013). Real-Time Dynamics of RNA Polymerase II Clustering in Live Human Cells. Science 341: 664-667.
Cook P. R. (1999). The Organization of Replication and Transcription. Science 284: 1790-1795.
Core L., Adelman K. (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes & Development 33: 960-982.
Core L. J., Waterfall J. J., Lis J. T. (2008). Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science 322: 1845-1848.
Corrigan A. M., Tunnacliffe E., Cannon D., Chubb J. R. (2016). A continuum model of transcriptional bursting. eLife 5: e13051.
Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., Jaenisch R. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences 107: 21931-21936.
Cruz-Molina S., Respuela P., Tebartz C., Kolovos P., Nikolic M., Fueyo R., van Ijcken W. F.J., Grosveld F., Frommolt P., Bazzi H., Rada-Iglesias A. (2017). PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell 20: 689-705.e9.
Cusanovich D. A., Reddington J. P., Garfield D. A., Daza R. M., Aghamirzaie D., Marco-Ferreres R., Pliner H. A., Christiansen L., Qiu X., Steemers F. J., Trapnell C., Shendure J., Furlong E. E. M. (2018). The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555: 538-542.
Dahl J. A., Jung I., Aanes H., Greggains G. D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A. Y., Preissl S., Jermstad I., Haugen M. H., Suganthan R., Bjørås M., Hansen K., Dalen K. T., Fedorcsak P., Ren B., Klungland A. (2016). Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537: 548-552.
Dancy B. M., Cole P. A. (2015). Protein Lysine Acetylation by p300/CBP. Chemical Reviews 115: 2419-2452.
Danko C. G., Hah N., Luo X., Martins A. L., Core L., Lis J. T., Siepel A., Kraus W. L. (2013). Signaling Pathways Differentially Affect RNA Polymerase II Initiation, Pausing, and Elongation Rate in Cells. Molecular Cell 50: 212-222.
Day D. S., Zhang B., Stevens S. M., Ferrari F., Larschan E. N., Park P. J., Pu W. T. (2016). Comprehensive analysis of promoter-proximal RNA polymerase II pausing across mammalian cell types. Genome Biology 17: 120.
Deng W., Lee J., Wang H., Miller J., Reik A., Gregory P. D., Dean A., Blobel G. A. (2012). Controlling Long-Range Genomic Interactions at a Native Locus by Targeted Tethering of a Looping Factor. Cell 149: 1233-1244.
Deng W., Rupon J. W., Krivega I., Breda L., Motta I., Jahn K. S., Reik A., Gregory P. D., Rivella S., Dean A., Blobel G. A. (2014). Reactivation of Developmentally Silenced Globin Genes by Forced Chromatin Looping. Cell 158: 849-860.
Dey A., Chitsaz F., Abbasi A., Misteli T., Ozato K. (2003). The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proceedings of the National Academy of Sciences 100: 8758-8763.
Dixon J. R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J. E., Lee A. Y., Ye Z., Kim A., Rajagopal N., Xie W., Diao Y., Liang J., Zhao H., Lobanenkov V. V., Ecker J. R., Thomson J. A., Ren B. (2015). Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331-336.
Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376-380.
Dorighi K. M., Swigut T., Henriques T., Bhanu N. V., Scruggs B. S., Nady N., Still C. D., Garcia B. A., Adelman K., Wysocka J. (2017). Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Molecular Cell 66: 568-576.e4.
Duan J., Rieder L., Colonnetta M. M., Huang A., Mckenney M., Watters S., Deshpande G., Jordan W., Fawzi N., Larschan E. (2021). CLAMP and Zelda function together to promote Drosophila zygotic genome activation. eLife 10: e69937.
Dubnicoff T., Valentine S. A., Chen G., Shi T., Lengyel J. A., Paroush Z., Courey A. J. (1997). Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes & Development 11: 2952-2957.
Dufourt J., Trullo A., Hunter J., Fernandez C., Lazaro J., Dejean M., Morales L., Nait-Amer S., Schulz K. N., Harrison M. M., Favard C., Radulescu O., Lagha M. (2018). Temporal control of gene expression by the pioneer factor Zelda through transient interactions in hubs. Nature Communications 9: 5194.
ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57-74.
Enderle D., Beisel C., Stadler M. B., Gerstung M., Athri P., Paro R. (2011). Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Research 21: 216-226.
Espinola S. M., Götz M., Bellec M., Messina O., Fiche J.B., Houbron C., Dejean M., Reim I., Cardozo Gizzi A. M., Lagha M., Nollmann M. (2021). Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nature Genetics 53: 477-486.
Feller C., Forné I., Imhof A., Becker P. B. (2015). Global and Specific Responses of the Histone Acetylome to Systematic Perturbation. Molecular Cell 57: 559-571.
Fenley A. T., Adams D. A., Onufriev A. V. (2010). Charge State of the Globular Histone Core Controls Stability of the Nucleosome. Biophysical Journal 99: 1577-1585.
Ferguson E. L., Anderson K. V. (1992). decapentaplegic acts as a morphogen to organize dorsal-ventral pattern in the Drosophila embryo. Cell 71: 451-461.
Fianu I., Chen Y., Dienemann C., Dybkov O., Linden A., Urlaub H., Cramer P. (2021). Structural basis of Integrator-mediated transcription regulation. Science 374: 883-887.
Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., Ward L. D., Brugman W., de Castro I. J., Kerkhoven R. M., Bussemaker H. J., van Steensel B. (2010). Systematic Protein Location Mapping Reveals Five Principal Chromatin Types in Drosophila Cells. Cell 143: 212-224.
Flavahan W. A., Gaskell E., Bernstein B. E. (2017). Epigenetic plasticity and the hallmarks of cancer. Science 357: aal2380.
Franke M., Ibrahim D. M., Andrey G., Schwarzer W., Heinrich V., Schöpflin R., Kraft K., Kempfer R., Jerković I., Chan W.L., Spielmann M., Timmermann B., Wittler L., Kurth I., Cambiaso P., Zuffardi O., Houge G., Lambie L., Brancati F., Pombo A., Vingron M., Spitz F., Mundlos S. (2016). Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265-269.
Frankel N., Davis G. K., Vargas D., Wang S., Payre F., Stern D. L. (2010). Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490-493.
Fujinaga K., Huang F., Peterlin B. M. (2023). P-TEFb: The master regulator of transcription elongation. Molecular Cell 83: 393-403.
Fujisawa T., Filippakopoulos P. (2017). Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nature Reviews Molecular Cell Biology 18: 246-262.
Fukaya T., Lim B., Levine M. (2016). Enhancer Control of Transcriptional Bursting. Cell 166: 358-368.
Furlong E. E. M., Levine M. (2018). Developmental enhancers and chromosome topology. Science 361: 1341-1345.
Gaertner B., Johnston J., Chen K., Wallaschek N., Paulson A., Garruss A. S., Gaudenz K., De Kumar B., Krumlauf R., Zeitlinger J. (2012). Poised RNA Polymerase II Changes over Developmental Time and Prepares Genes for Future Expression. Cell Reports 2: 1670-1683.
Gaertner B., Zeitlinger J. (2014). RNA polymerase II pausing during development. Development 141: 1179-1183.
Gaskill M. M., Gibson T. J., Larson E. D., Harrison M. M. (2021). GAF is essential for zygotic genome activation and chromatin accessibility in the early Drosophila embryo. eLife 10: e66668.
Ghavi-Helm Y., Jankowski A., Meiers S., Viales R. R., Korbel J. O., Furlong E. E. M. (2019). Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nature Genetics 51: 1272-1282.
Ghavi-Helm Y., Klein F. A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E. E. M. (2014). Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512: 96-100.
Gilchrist D. A., Dos Santos G., Fargo D. C., Xie B., Gao Y., Li L., Adelman K. (2010). Pausing of RNA Polymerase II Disrupts DNA-Specified Nucleosome Organization to Enable Precise Gene Regulation. Cell 143: 540-551.
Gillespie M. A., Palii C. G., Sanchez-Taltavull D., Shannon P., Longabaugh W. J.R., Downes D. J., Sivaraman K., Espinoza H. M., Hughes J. R., Price N. D., Perkins T. J., Ranish J. A., Brand M. (2020). Absolute Quantification of Transcription Factors Reveals Principles of Gene Regulation in Erythropoiesis. Molecular Cell 78: 960-974.e11.
Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. (1983). A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell 33: 717-728.
Gilmour D. S., Lis J. T. (1986). RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells.. Molecular and Cellular Biology 6: 3984-3989.
Glozak M. A., Sengupta N., Zhang X., Seto E. (2005). Acetylation and deacetylation of non-histone proteins. Gene 363: 15-23.
Guo Y. E., Manteiga J. C., Henninger J. E., Sabari B. R., Dall’Agnese A., Hannett N. M., Spille J.H., Afeyan L. K., Zamudio A. V., Shrinivas K., Abraham B. J., Boija A., Decker T.M., Rimel J. K., Fant C. B., Lee T. I., Cisse I. I., Sharp P. A., Taatjes D. J., Young R. A. (2019). Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572: 543-548.
Haberle V., Arnold C. D., Pagani M., Rath M., Schernhuber K., Stark A. (2019). Transcriptional cofactors display specificity for distinct types of core promoters. Nature 570: 122-126.
Haberle V., Stark A. (2018). Eukaryotic core promoters and the functional basis of transcription initiation. Nature Reviews Molecular Cell Biology 19: 621-637.
Hamaratoglu F., Affolter M., Pyrowolakis G. (2014). Dpp/BMP signaling in flies: From molecules to biology. Seminars in Cell & Developmental Biology 32: 128-136.
Harrison M. M., Eisen M. B. (2015). Transcriptional Activation of the Zygotic Genome in Drosophila. In The Maternal-to-Zygotic Transition. Elsevier.
Harrison M. M., Li X.Y., Kaplan T., Botchan M. R., Eisen M. B. (2011). Zelda Binding in the Early Drosophila melanogaster Embryo Marks Regions Subsequently Activated at the Maternal-to-Zygotic Transition. PLoS Genetics 7: e1002266.
Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., Lobanenkov V. V., Stewart R., Thomson J. A., Crawford G. E., Kellis M., Ren B. (2009). Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108-112.
Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics 39: 311-318.
Heist T., Fukaya T., Levine M. (2019). Large distances separate coregulated genes in living Drosophila embryos. Proceedings of the National Academy of Sciences 116: 15062-15067.
Hendy O., Serebreni L., Bergauer K., Muerdter F., Huber L., Nemčko F., Stark A. (2022). Developmental and housekeeping transcriptional programs in Drosophila require distinct chromatin remodelers. Molecular Cell 82: 3598-3612.e7.
Henikoff S., Shilatifard A. (2011). Histone modification: cause or cog?. Trends in Genetics 27: 389-396.
Henninger J. E., Oksuz O., Shrinivas K., Sagi I., LeRoy G., Zheng M. M., Andrews J. O., Zamudio A. V., Lazaris C., Hannett N. M., Lee T. I., Sharp P. A., Cissé I. I., Chakraborty A. K., Young R. A. (2021). RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 184: 207-225.e24.
Henriques T., Gilchrist D. A., Nechaev S., Bern M., Muse G. W., Burkholder A., Fargo D. C., Adelman K. (2013). Stable Pausing by RNA Polymerase II Provides an Opportunity to Target and Integrate Regulatory Signals. Molecular Cell 52: 517-528.
Hnisz D., Shrinivas K., Young R. A., Chakraborty A. K., Sharp P. A. (2017). A Phase Separation Model for Transcriptional Control. Cell 169: 13-23.
Hogg S. J., Motorna O., Cluse L. A., Johanson T. M., Coughlan H. D., Raviram R., Myers R. M., Costacurta M., Todorovski I., Pijpers L., Bjelosevic S., Williams T., Huskins S. N., Kearney C. J., Devlin J. R., Fan Z., Jabbari J. S., Martin B. P., Fareh M., Kelly M. J., Dupéré-Richer D., Sandow J. J., Feran B., Knight D., Khong T., Spencer A., Harrison S. J., Gregory G., Wickramasinghe V. O., Webb A. I., Taberlay P. C., Bromberg K. D., Lai A., Papenfuss A. T., Smyth G. K., Allan R. S., Licht J. D., Landau D. A., Abdel-Wahab O., Shortt J., Vervoort S. J., Johnstone R. W. (2021). Targeting histone acetylation dynamics and oncogenic transcription by catalytic P300/CBP inhibition. Molecular Cell 81: 2183-2200.e13.
Holmqvist P.H., Boija A., Philip P., Crona F., Stenberg P., Mannervik M. (2012). Preferential Genome Targeting of the CBP Co-Activator by Rel and Smad Proteins in Early Drosophila melanogaster Embryos. PLoS Genetics 8: e1002769.
Hong J.W., Hendrix D. A., Levine M. S. (2008a). Shadow Enhancers as a Source of Evolutionary Novelty. Science 321: 1314-1314.
Hong J.W., Hendrix D. A., Papatsenko D., Levine M. S. (2008b). How the Dorsal gradient works: Insights from postgenome technologies. Proceedings of the National Academy of Sciences 105: 20072-20076.
Hornung G., Bar-Ziv R., Rosin D., Tokuriki N., Tawfik D. S., Oren M., Barkai N. (2012). Noise–mean relationship in mutated promoters. Genome Research 22: 2409-2417.
Hou C., Li L., Qin Z. S., Corces V. G. (2012). Gene Density, Transcription, and Insulators Contribute to the Partition of the Drosophila Genome into Physical Domains. Molecular Cell 48: 471-484.
Hsieh T.H. S., Cattoglio C., Slobodyanyuk E., Hansen A. S., Darzacq X., Tjian R. (2022). Enhancer–promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nature Genetics 54: 1919-1932.
Hsieh T.H. S., Cattoglio C., Slobodyanyuk E., Hansen A. S., Rando O. J., Tjian R., Darzacq X. (2020). Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Molecular Cell 78: 539-553.e8.
Huang A., Amourda C., Zhang S., Tolwinski N. S., Saunders T. E. (2017). Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. eLife 6: e26258.
Hug C. B., Grimaldi A. G., Kruse K., Vaquerizas J. M. (2017). Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 169: 216-228.e19.
Hunt G., Boija A., Mannervik M. (2022). p300/CBP sustains Polycomb silencing by non-enzymatic functions. Molecular Cell 82: 3580-3597.e9.
Hunt G., Vaid R., Pirogov S., Pfab A., Ziegenhain C., Sandberg R., Reimegård J., Mannervik M. (2024). Tissue-specific RNA Polymerase II promoter-proximal pause release and burst kinetics in a Drosophila embryonic patterning network. Genome Biology 25: 2.
Iborra F. J., Pombo A., Jackson D. A., Cook P. R. (1996). Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. Journal of Cell Science 109: 1427-1436.
Ing-Simmons E., Vaid R., Bing X. Y., Levine M., Mannervik M., Vaquerizas J. M. (2021). Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nature Genetics 53: 487-499.
Irizarry J., Stathopoulos A. (2021). Dynamic patterning by morphogens illuminated by cis-regulatory studies. Development 148: dev196113.
Jang M. K., Mochizuki K., Zhou M., Jeong H.S., Brady J. N., Ozato K. (2005). The Bromodomain Protein Brd4 Is a Positive Regulatory Component of P-TEFb and Stimulates RNA Polymerase II-Dependent Transcription. Molecular Cell 19: 523-534.
Jerkovic I., Cavalli G. (2021). Understanding 3D genome organization by multidisciplinary methods. Nature Reviews Molecular Cell Biology 22: 511-528.
Jiang N., Emberly E., Cuvier O., Hart C. M. (2009). Genome-Wide Mapping of Boundary Element-Associated Factor (BEAF) Binding Sites in Drosophila melanogaster Links BEAF to Transcription. Molecular and Cellular Biology 29: 3556-3568.
Jiang Y., Zhang N. R., Li M. (2017). SCALE: modeling allele-specific gene expression by single-cell RNA sequencing. Genome Biology 18: 74.
Jin F., Li Y., Dixon J. R., Selvaraj S., Ye Z., Lee A. Y., Yen C.A., Schmitt A. D., Espinoza C. A., Ren B. (2013). A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503: 290-294.
Jonkers I., Kwak H., Lis J. T. (2014). Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3: e02407.
Jonkers I., Lis J. T. (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nature Reviews Molecular Cell Biology 16: 167-177.
Joo Y. J., Ficarro S. B., Soares L. M., Chun Y., Marto J. A., Buratowski S. (2017). Downstream promoter interactions of TFIID TAFs facilitate transcription reinitiation. Genes & Development 31: 2162-2174.
Kanhere A., Viiri K., Araújo C. C., Rasaiyaah J., Bouwman R. D., Whyte W. A., Pereira C. F., Brookes E., Walker K., Bell G. W., Pombo A., Fisher A. G., Young R. A., Jenner R. G. (2010). Short RNAs Are Transcribed from Repressed Polycomb Target Genes and Interact with Polycomb Repressive Complex-2. Molecular Cell 38: 675-688.
Karaiskos N., Wahle P., Alles J., Boltengagen A., Ayoub S., Kipar C., Kocks C., Rajewsky N., Zinzen R. P. (2017). The Drosophila embryo at single-cell transcriptome resolution. Science 358: 194-199.
Karr J. P., Ferrie J. J., Tjian R., Darzacq X. (2022). The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer–promoter communication. Genes & Development 36: 7-16.
Kaushal A., Dorier J., Wang B., Mohana G., Taschner M., Cousin P., Waridel P., Iseli C., Semenova A., Restrepo S., Guex N., Aiden E. L., Gambetta M. C. (2022). Essential role of Cp190 in physical and regulatory boundary formation. Science Advances 8: eabl8834.
Kaushal A., Mohana G., Dorier J., Özdemir I., Omer A., Cousin P., Semenova A., Taschner M., Dergai O., Marzetta F., Iseli C., Eliaz Y., Weisz D., Shamim M. S., Guex N., Lieberman Aiden E., Gambetta M. C. (2021). CTCF loss has limited effects on global genome architecture in Drosophila despite critical regulatory functions. Nature Communications 12: 1011.
Kawasaki K., Fukaya T. (2023). Functional coordination between transcription factor clustering and gene activity. Molecular Cell 83: 1605-1622.e9.
Kim J., Marioni J. C. (2013). Inferring the kinetics of stochastic gene expression from single-cell RNA-sequencing data. Genome Biology 14: R7.
Koenecke N., Johnston J., Gaertner B., Natarajan M., Zeitlinger J. (2016). Genome-wide identification of Drosophila dorso-ventral enhancers by differential histone acetylation analysis. Genome Biology 17: 196.
Koenecke N., Johnston J., He Q., Meier S., Zeitlinger J. (2017). Drosophila poised enhancers are generated during tissue patterning with the help of repression. Genome Research 27: 64-74.
Kornberg R. D., Lorch Y. (2020). Primary Role of the Nucleosome. Molecular Cell 79: 371-375.
Koromila T., Gao F., Iwasaki Y., He P., Pachter L., Gergen J. P., Stathopoulos A. (2020). Odd-paired is a pioneer-like factor that coordinates with Zelda to control gene expression in embryos. eLife 9: e59610.
Kosman D., Ip Y. T., Levine M., Arora K. (1991). Establishment of the Mesoderm-Neuroectoderm Boundary in the Drosophila Embryo. Science 254: 118-122.
Kragesteen B. K., Spielmann M., Paliou C., Heinrich V., Schöpflin R., Esposito A., Annunziatella C., Bianco S., Chiariello A. M., Jerković I., Harabula I., Guckelberger P., Pechstein M., Wittler L., Chan W.L., Franke M., Lupiáñez D. G., Kraft K., Timmermann B., Vingron M., Visel A., Nicodemi M., Mundlos S., Andrey G. (2018). Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nature Genetics 50: 1463-1473.
Krietenstein N., Abraham S., Venev S. V., Abdennur N., Gibcus J., Hsieh T.H. S., Parsi K. M., Yang L., Maehr R., Mirny L. A., Dekker J., Rando O. J. (2020). Ultrastructural Details of Mammalian Chromosome Architecture. Molecular Cell 78: 554-565.e7.
Kvon E. Z., Kazmar T., Stampfel G., Yáñez-Cuna J. O., Pagani M., Schernhuber K., Dickson B. J., Stark A. (2014). Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature 512: 91-95.
Kwok R. P. S., Lundblad J. R., Chrivia J. C., Richards J. P., Bächinger H. P., Brennan R. G., Roberts S. G. E., Green M. R., Goodman R. H. (1994). Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370: 223-226.
Lagha M., Bothma J. P., Esposito E., Ng S., Stefanik L., Tsui C., Johnston J., Chen K., Gilmour D. S., Zeitlinger J., Levine M. S. (2013). Paused Pol II Coordinates Tissue Morphogenesis in the Drosophila Embryo. Cell 153: 976-987.
Larsson A. J. M., Johnsson P., Hagemann-Jensen M., Hartmanis L., Faridani O. R., Reinius B., Segerstolpe Å, Rivera C. M., Ren B., Sandberg R. (2019). Genomic encoding of transcriptional burst kinetics. Nature 565: 251-254.
Le Dily F., Baù D., Pohl A., Vicent G. P., Serra F., Soronellas D., Castellano G., Wright R. H.G., Ballare C., Filion G., Marti-Renom M. A., Beato M. (2014). Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes & Development 28: 2151-2162.
Leatham-Jensen M., Uyehara C. M., Strahl B. D., Matera A. G., Duronio R. J., McKay D. J. (2019). Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation. PLOS Genetics 15: e1007932.
Levine M. (2010). Transcriptional Enhancers in Animal Development and Evolution. Current Biology 20: R754-R763.
Levine M. (2011). Paused RNA Polymerase II as a Developmental Checkpoint. Cell 145: 502-511.
Levo M., Raimundo J., Bing X. Y., Sisco Z., Batut P. J., Ryabichko S., Gregor T., Levine M. S. (2022). Transcriptional coupling of distant regulatory genes in living embryos. Nature 605: 754-760.
Leyes Porello E. A., Trudeau R. T., Lim B. (2023). Transcriptional bursting: stochasticity in deterministic development. Development 150: dev201546.
Li B., Carey M., Workman J. L. (2007). The Role of Chromatin during Transcription. Cell 128: 707-719.
Li G., Ruan X., Auerbach R. K., Sandhu K. S., Zheng M., Wang P., Poh H. M., Goh Y., Lim J., Zhang J., Sim H. S., Peh S. Q., Mulawadi F. H., Ong C. T., Orlov Y. L., Hong S., Zhang Z., Landt S., Raha D., Euskirchen G., Wei C.L., Ge W., Wang H., Davis C., Fisher-Aylor K. I., Mortazavi A., Gerstein M., Gingeras T., Wold B., Sun Y., Fullwood M. J., Cheung E., Liu E., Sung W.K., Snyder M., Ruan Y. (2012). Extensive Promoter-Centered Chromatin Interactions Provide a Topological Basis for Transcription Regulation. Cell 148: 84-98.
Li X., Tang X., Bing X., Catalano C., Li T., Dolsten G., Wu C., Levine M. (2023). GAGA-associated factor fosters loop formation in the Drosophila genome. Molecular Cell 83: 1519-1526.e4.
Li X.Y., Harrison M. M., Villalta J. E., Kaplan T., Eisen M. B. (2014). Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 3: e03737.
Li X.Y., Thomas S., Sabo P. J., Eisen M. B., Stamatoyannopoulos J. A., Biggin M. D. (2011). The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome Biology 12: R34.
Liang H.L., Nien C.Y., Liu H.Y., Metzstein M. M., Kirov N., Rushlow C. (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400-403.
Liberman L. M., Reeves G. T., Stathopoulos A. (2009). Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila. Proceedings of the National Academy of Sciences 106: 22317-22322.
Lim B., Heist T., Levine M., Fukaya T. (2018). Visualization of Transvection in Living Drosophila Embryos. Molecular Cell 70: 287-296.e6.
Lim B., Levine M. S. (2021). Enhancer-promoter communication: hubs or loops?. Current Opinion in Genetics & Development 67: 5-9.
Liu B., Xu Q., Wang Q., Feng S., Lai F., Wang P., Zheng F., Xiang Y., Wu J., Nie J., Qiu C., Xia W., Li L., Yu G., Lin Z., Xu K., Xiong Z., Kong F., Liu L., Huang C., Yu Y., Na J., Xie W. (2020). The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 587: 139-144.
Long H. K., Prescott S. L., Wysocka J. (2016). Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 167: 1170-1187.
López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G. (2013). The Hallmarks of Aging. Cell 153: 1194-1217.
Lott S. E., Villalta J. E., Schroth G. P., Luo S., Tonkin L. A., Eisen M. B. (2011). Noncanonical Compensation of Zygotic X Transcription in Early Drosophila melanogaster Development Revealed through Single-Embryo RNA-Seq. PLoS Biology 9: e1000590.
Lu H., Yu D., Hansen A. S., Ganguly S., Liu R., Heckert A., Darzacq X., Zhou Q. (2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558: 318-323.
Lupiáñez D. G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J. M., Laxova R., Santos-Simarro F., Gilbert-Dussardier B., Wittler L., Borschiwer M., Haas S. A., Osterwalder M., Franke M., Timmermann B., Hecht J., Spielmann M., Visel A., Mundlos S. (2015). Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 161: 1012-1025.
Ma J., He F., Xie G., Deng W.M. (2016). Maternal AP determinants in the Drosophila oocyte and embryo. WIREs Developmental Biology 5: 562-581.
Ma L., Gao Z., Wu J., Zhong B., Xie Y., Huang W., Lin Y. (2021). Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Molecular Cell 81: 1682-1697.e7.
Mannervik M., Nibu Y., Zhang H., Levine M. (1999). Transcriptional Coregulators in Development. Science 284: 606-609.
Markstein M., Markstein P., Markstein V., Levine M. S. (2002). Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proceedings of the National Academy of Sciences 99: 763-768.
Mateo L. J., Murphy S. E., Hafner A., Cinquini I. S., Walker C. A., Boettiger A. N. (2019). Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49-54.
McDaniel S. L., Gibson T. J., Schulz K. N., Fernandez Garcia M., Nevil M., Jain S. U., Lewis P. W., Zaret K. S., Harrison M. M. (2019). Continued Activity of the Pioneer Factor Zelda Is Required to Drive Zygotic Genome Activation. Molecular Cell 74: 185-195.e4.
McKay D. J., Klusza S., Penke T. J.R., Meers M. P., Curry K. P., McDaniel S. L., Malek P. Y., Cooper S. W., Tatomer D. C., Lieb J. D., Strahl B. D., Duronio R. J., Matera A. G. (2015). Interrogating the Function of Metazoan Histones using Engineered Gene Clusters. Developmental Cell 32: 373-386.
McSwiggen D. T., Mir M., Darzacq X., Tjian R. (2019). Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes & Development 33: 1619-1634.
Miao L., Tang Y., Bonneau A. R., Chan S. H., Kojima M. L., Pownall M. E., Vejnar C. E., Gao F., Krishnaswamy S., Hendry C. E., Giraldez A. J. (2022). The landscape of pioneer factor activity reveals the mechanisms of chromatin reprogramming and genome activation. Molecular Cell 82: 986-1002.e9.
Millán-Zambrano G., Burton A., Bannister A. J., Schneider R. (2022). Histone post-translational modifications — cause and consequence of genome function. Nature Reviews Genetics 23: 563-580.
Miller Jr. O. L., McKnight S. L. (1979). Post-replicative nonribosomal transcription units in D. melanogaster embryos. Cell 17: 551-563.
Mir M., Stadler M. R., Ortiz S. A., Hannon C. E., Harrison M. M., Darzacq X., Eisen M. B. (2018). Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 7: e40497.
Mito Y., Henikoff J. G., Henikoff S. (2007). Histone Replacement Marks the Boundaries of cis-Regulatory Domains. Science 315: 1408-1411.
Morgan M. A. J., Shilatifard A. (2020). Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nature Genetics 52: 1271-1281.
Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., Adelman K. (2007). RNA polymerase is poised for activation across the genome. Nature Genetics 39: 1507-1511.
Narita T., Higashijima Y., Kilic S., Liebner T., Walter J., Choudhary C. (2023). Acetylation of histone H2B marks active enhancers and predicts CBP/p300 target genes. Nature Genetics 55: 679-692.
Narita T., Ito S., Higashijima Y., Chu W. K., Neumann K., Walter J., Satpathy S., Liebner T., Hamilton W. B., Maskey E., Prus G., Shibata M., Iesmantavicius V., Brickman J. M., Anastassiadis K., Koseki H., Choudhary C. (2021). Enhancers are activated by p300/CBP activity-dependent PIC assembly, RNAPII recruitment, and pause release. Molecular Cell 81: 2166-2182.e6.
Nechaev S., Fargo D. C., dos Santos G., Liu L., Gao Y., Adelman K. (2010). Global Analysis of Short RNAs Reveals Widespread Promoter-Proximal Stalling and Arrest of Pol II in Drosophila. Science 327: 335-338.
Nègre N., Brown C. D., Ma L., Bristow C. A., Miller S. W., Wagner U., Kheradpour P., Eaton M. L., Loriaux P., Sealfon R., Li Z., Ishii H., Spokony R. F., Chen J., Hwang L., Cheng C., Auburn R. P., Davis M. B., Domanus M., Shah P. K., Morrison C. A., Zieba J., Suchy S., Senderowicz L., Victorsen A., Bild N. A., Grundstad A. J., Hanley D., MacAlpine D. M., Mannervik M., Venken K., Bellen H., White R., Gerstein M., Russell S., Grossman R. L., Ren B., Posakony J. W., Kellis M., White K. P. (2011). A cis-regulatory map of the Drosophila genome. Nature 471: 527-531.
Neuert G., Munsky B., Tan R. Z., Teytelman L., Khammash M., van Oudenaarden A. (2013). Systematic Identification of Signal-Activated Stochastic Gene Regulation. Science 339: 584-587.
Neumayr C., Haberle V., Serebreni L., Karner K., Hendy O., Boija A., Henninger J. E., Li C. H., Stejskal K., Lin G., Bergauer K., Pagani M., Rath M., Mechtler K., Arnold C. D., Stark A. (2022). Differential cofactor dependencies define distinct types of human enhancers. Nature 606: 406-413.
Nibu Y., Zhang H., Levine M. (1998). Interaction of Short-Range Repressors with Drosophila CtBP in the Embryo. Science 280: 101-104.
Nicolas D., Zoller B., Suter D. M., Naef F. (2018). Modulation of transcriptional burst frequency by histone acetylation. Proceedings of the National Academy of Sciences 115: 7153-7158.
Nien C.Y., Liang H.L., Butcher S., Sun Y., Fu S., Gocha T., Kirov N., Manak J. R., Rushlow C. (2011). Temporal Coordination of Gene Networks by Zelda in the Early Drosophila Embryo. PLoS Genetics 7: e1002339.
Nora E. P., Goloborodko A., Valton A.L., Gibcus J. H., Uebersohn A., Abdennur N., Dekker J., Mirny L. A., Bruneau B. G. (2017). Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169: 930-944.e22.
Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N. L., Meisig J., Sedat J., Gribnau J., Barillot E., Blüthgen N., Dekker J., Heard E. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381-385.
Ogiyama Y., Schuettengruber B., Papadopoulos G. L., Chang J.M., Cavalli G. (2018). Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Molecular Cell 71: 73-88.e5.
Ortega E., Rengachari S., Ibrahim Z., Hoghoughi N., Gaucher J., Holehouse A. S., Khochbin S., Panne D. (2018). Transcription factor dimerization activates the p300 acetyltransferase. Nature 562: 538-544.
Osterwalder M., Barozzi I., Tissières V., Fukuda-Yuzawa Y., Mannion B. J., Afzal S. Y., Lee E. A., Zhu Y., Plajzer-Frick I., Pickle C. S., Kato M., Garvin T. H., Pham Q. T., Harrington A. N., Akiyama J. A., Afzal V., Lopez-Rios J., Dickel D. E., Visel A., Pennacchio L. A. (2018). Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554: 239-243.
Panigrahi A., O’Malley B. W. (2021). Mechanisms of enhancer action: the known and the unknown. Genome Biology 22: 108.
Papagianni A., Forés M., Shao W., He S., Koenecke N., Andreu M. J., Samper N., Paroush Z., González-Crespo S., Zeitlinger J., Jiménez G. (2018). Capicua controls Toll/IL-1 signaling targets independently of RTK regulation. Proceedings of the National Academy of Sciences 115: 1807-1812.
Papatsenko D., Levine M. (2005). Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proceedings of the National Academy of Sciences 102: 4966-4971.
Peccoud J., Ycart B. (1995). Markovian Modeling of Gene-Product Synthesis. Theoretical Population Biology 48: 222-234.
Pengelly A. R., Copur Ö., Jäckle H., Herzig A., Müller J. (2013). A Histone Mutant Reproduces the Phenotype Caused by Loss of Histone-Modifying Factor Polycomb. Science 339: 698-699.
Pennacchio L. A., Ahituv N., Moses A. M., Prabhakar S., Nobrega M. A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K. D., Plajzer-Frick I., Akiyama J., De Val S., Afzal V., Black B. L., Couronne O., Eisen M. B., Visel A., Rubin E. M. (2006). In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499-502.
Perry M. W., Boettiger A. N., Bothma J. P., Levine M. (2010). Shadow Enhancers Foster Robustness of Drosophila Gastrulation. Current Biology 20: 1562-1567.
Philip P., Boija A., Vaid R., Churcher A. M., Meyers D. J., Cole P. A., Mannervik M., Stenberg P. (2015). CBP binding outside of promoters and enhancers in Drosophila melanogaster. Epigenetics & Chromatin 8: 48.
Pimmett V. L., Dejean M., Fernandez C., Trullo A., Bertrand E., Radulescu O., Lagha M. (2021). Quantitative imaging of transcription in living Drosophila embryos reveals the impact of core promoter motifs on promoter state dynamics. Nature Communications 12: 4504.
Pique-Regi R., Degner J. F., Pai A. A., Gaffney D. J., Gilad Y., Pritchard J. K. (2011). Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Research 21: 447-455.
Pownall M. E., Miao L., Vejnar C. E., M’Saad O., Sherrard A., Frederick M. A., Benitez M. D. J., Boswell C. W., Zaret K. S., Bewersdorf J., Giraldez A. J. (2023). Chromatin expansion microscopy reveals nanoscale organization of transcription and chromatin. Science 381: 92-100.
Price D. H. (2000). P-TEFb, a Cyclin-Dependent Kinase Controlling Elongation by RNA Polymerase II. Molecular and Cellular Biology 20: 2629-2634.
Qi D., Bergman M., Aihara H., Nibu Y., Mannervik M. (2008). Drosophila Ebi mediates Snail-dependent transcriptional repression through HDAC3-induced histone deacetylation. The EMBO Journal 27: 898-909.
Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279-283.
Raj A., Peskin C. S., Tranchina D., Vargas D. Y., Tyagi S. (2006). Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biology 4: e309.
Ramalingam V., Natarajan M., Johnston J., Zeitlinger J. (2021). TATA and paused promoters active in differentiated tissues have distinct expression characteristics. Molecular Systems Biology 17: e9866.
Rao S. S.P., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., Robinson J. T., Sanborn A. L., Machol I., Omer A. D., Lander E. S., Aiden E. L. (2014). A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell 159: 1665-1680.
Rao S. S.P., Huang S.C., Glenn St Hilaire B., Engreitz J. M., Perez E. M., Kieffer-Kwon K.R., Sanborn A. L., Johnstone S. E., Bascom G. D., Bochkov I. D., Huang X., Shamim M. S., Shin J., Turner D., Ye Z., Omer A. D., Robinson J. T., Schlick T., Bernstein B. E., Casellas R., Lander E. S., Aiden E. L. (2017). Cohesin Loss Eliminates All Loop Domains. Cell 171: 305-320.e24.
Reddington J. P., Garfield D. A., Sigalova O. M., Karabacak Calviello A., Marco-Ferreres R., Girardot C., Viales R. R., Degner J. F., Ohler U., Furlong E. E.M. (2020). Lineage-Resolved Enhancer and Promoter Usage during a Time Course of Embryogenesis. Developmental Cell 55: 648-664.e9.
Reeves G. T., Stathopoulos A. (2009). Graded Dorsal and Differential Gene Regulation in the Drosophila Embryo. Cold Spring Harbor Perspectives in Biology 1: a000836-a000836.
Reeves G. T., Trisnadi N., Truong T. V., Nahmad M., Katz S., Stathopoulos A. (2012). Dorsal-Ventral Gene Expression in the Drosophila Embryo Reflects the Dynamics and Precision of the Dorsal Nuclear Gradient. Developmental Cell 22: 544-557.
Reiter F., Wienerroither S., Stark A. (2017). Combinatorial function of transcription factors and cofactors. Current Opinion in Genetics & Development 43: 73-81.
Rickels R., Herz H.M., Sze C. C., Cao K., Morgan M. A., Collings C. K., Gause M., Takahashi Y., Wang L., Rendleman E. J., Marshall S. A., Krueger A., Bartom E. T., Piunti A., Smith E. R., Abshiru N. A., Kelleher N. L., Dorsett D., Shilatifard A. (2017). Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nature Genetics 49: 1647-1653.
Rickels R., Shilatifard A. (2018). Enhancer Logic and Mechanics in Development and Disease. Trends in Cell Biology 28: 608-630.
Rincon-Arano H., Halow J., Delrow J. J., Parkhurst S. M., Groudine M. (2012). UpSET Recruits HDAC Complexes and Restricts Chromatin Accessibility and Acetylation at Promoter Regions. Cell 151: 1214-1228.
Rodriguez J., Larson D. R. (2020). Transcription in Living Cells: Molecular Mechanisms of Bursting. Annual Review of Biochemistry 89: 189-212.
Roth S., Stein D., Nüsslein-Volhard C. (1989). A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59: 1189-1202.
Rougvie A. E., Lis J. T. (1988). The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795-804.
Rusch J., Levine M. (1996). Threshold responses to the dorsal regulatory gradient and the subdivision of primary tissue territories in the Drosophila embryo. Current Opinion in Genetics & Development 6: 416-423.
Rushlow C. A., Han K., Manley J. L., Levine M. (1989). The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell 59: 1165-1177.
Sabari B. R., Dall’Agnese A., Boija A., Klein I. A., Coffey E. L., Shrinivas K., Abraham B. J., Hannett N. M., Zamudio A. V., Manteiga J. C., Li C. H., Guo Y. E., Day D. S., Schuijers J., Vasile E., Malik S., Hnisz D., Lee T. I., Cisse I. I., Roeder R. G., Sharp P. A., Chakraborty A. K., Young R. A. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361: eaar3958.
Sankar A., Mohammad F., Sundaramurthy A. K., Wang H., Lerdrup M., Tatar T., Helin K. (2022). Histone editing elucidates the functional roles of H3K27 methylation and acetylation in mammals. Nature Genetics 54: 754-760.
Sanyal A., Lajoie B. R., Jain G., Dekker J. (2012). The long-range interaction landscape of gene promoters. Nature 489: 109-113.
Sato Y., Hilbert L., Oda H., Wan Y., Heddleston J. M., Chew T.L., Zaburdaev V., Keller P., Lionnet T., Vastenhouw N., Kimura H. (2019). Histone H3K27 acetylation precedes active transcription during zebrafish zygotic genome activation as revealed by live-cell analysis. Development 146: dev179127.
Saunders A., Core L. J., Sutcliffe C., Lis J. T., Ashe H. L. (2013). Extensive polymerase pausing during Drosophila axis patterning enables high-level and pliable transcription. Genes & Development 27: 1146-1158.
Schoenfelder S., Fraser P. (2019). Long-range enhancer–promoter contacts in gene expression control. Nature Reviews Genetics 20: 437-455.
Schröder S., Herker E., Itzen F., He D., Thomas S., Gilchrist D. A., Kaehlcke K., Cho S., Pollard K. S., Capra J. A., Schnölzer M., Cole P. A., Geyer M., Bruneau B. G., Adelman K., Ott M. (2013). Acetylation of RNA Polymerase II Regulates Growth-Factor-Induced Gene Transcription in Mammalian Cells. Molecular Cell 52: 314-324.
Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. (2017). Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171: 34-57.
Schuettengruber B., Ganapathi M., Leblanc B., Portoso M., Jaschek R., Tolhuis B., van Lohuizen M., Tanay A., Cavalli G. (2009). Functional Anatomy of Polycomb and Trithorax Chromatin Landscapes in Drosophila Embryos. PLoS Biology 7: e1000013.
Schulz K. N., Harrison M. M. (2019). Mechanisms regulating zygotic genome activation. Nature Reviews Genetics 20: 221-234.
Schwartz Y. B., Kahn T. G., Nix D. A., Li X.Y., Bourgon R., Biggin M., Pirrotta V. (2006). Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nature Genetics 38: 700-705.
Schwarzer W., Abdennur N., Goloborodko A., Pekowska A., Fudenberg G., Loe-Mie Y., Fonseca N. A., Huber W., Haering C. H., Mirny L., Spitz F. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551: 51-56.
Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. (2012). Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 148: 458-472.
Shao W., Zeitlinger J. (2017). Paused RNA polymerase II inhibits new transcriptional initiation. Nature Genetics 49: 1045-1051.
Shlyueva D., Stampfel G., Stark A. (2014). Transcriptional enhancers: from properties to genome-wide predictions. Nature Reviews Genetics 15: 272-286.
Shrinivas K., Sabari B. R., Coffey E. L., Klein I. A., Boija A., Zamudio A. V., Schuijers J., Hannett N. M., Sharp P. A., Young R. A., Chakraborty A. K. (2019). Enhancer Features that Drive Formation of Transcriptional Condensates. Molecular Cell 75: 549-561.e7.
Singh A. P., Wu P., Ryabichko S., Raimundo J., Swan M., Wieschaus E., Gregor T., Toettcher J. E. (2022). Optogenetic control of the Bicoid morphogen reveals fast and slow modes of gap gene regulation. Cell Reports 38: 110543.
Sinha K. K., Bilokapic S., Du Y., Malik D., Halic M. (2023). Histone modifications regulate pioneer transcription factor cooperativity. Nature 619: 378-384.
Small S., Arnosti D. N. (2020). Transcriptional Enhancers in Drosophila. Genetics 216: 1-26.
Small S., Blair A., Levine M. (1992). Regulation of even-skipped stripe 2 in the Drosophila embryo.. The EMBO Journal 11: 4047-4057.
Smith G. D., Ching W. H., Cornejo-Páramo P., Wong E. S. (2023). Decoding enhancer complexity with machine learning and high-throughput discovery. Genome Biology 24: 116.
Soluri I. V., Zumerling L. M., Payan Parra O. A., Clark E. G., Blythe S. A. (2020). Zygotic pioneer factor activity of Odd-paired/Zic is necessary for late function of the Drosophila segmentation network. eLife 9: e53916.
Spange S., Wagner T., Heinzel T., Krämer O. H. (2009). Acetylation of non-histone proteins modulates cellular signalling at multiple levels. The International Journal of Biochemistry & Cell Biology 41: 185-198.
Spitz F., Furlong E. E. M. (2012). Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics 13: 613-626.
Stadler M. R., Haines J. E., Eisen M. B. (2017). Convergence of topological domain boundaries, insulators, and polytene interbands revealed by high-resolution mapping of chromatin contacts in the early Drosophila melanogaster embryo. eLife 6: e29550.
Stanojevic D., Small S., Levine M. (1991). Regulation of a Segmentation Stripe by Overlapping Activators and Repressors in the Drosophila Embryo. Science 254: 1385-1387.
Stasevich T. J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., Sakata-Sogawa K., Tokunaga M., Nagase T., Nozaki N., McNally J. G., Kimura H. (2014). Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 516: 272-275.
Stathopoulos A., Levine M. (2002). Dorsal Gradient Networks in the Drosophila Embryo. Developmental Biology 246: 57-67.
Stathopoulos A., Levine M. (2004). Whole-genome analysis of Drosophila gastrulation. Current Opinion in Genetics & Development 14: 477-484.
Stathopoulos A., Van Drenth M., Erives A., Markstein M., Levine M. (2002). Whole-Genome Analysis of Dorsal-Ventral Patterning in the Drosophila Embryo. Cell 111: 687-701.
Stein C. B., Field A. R., Mimoso C. A., Zhao C.C., Huang K.L., Wagner E. J., Adelman K. (2022). Integrator endonuclease drives promoter-proximal termination at all RNA polymerase II-transcribed loci. Molecular Cell 82: 4232-4245.e11.
Stein D. S., Stevens L. M. (2014). Maternal control of the Drosophila dorsal–ventral body axis. WIREs Developmental Biology 3: 301-330.
Steward R. (1987). Dorsal , an Embryonic Polarity Gene in Drosophila , Is Homologous to the Vertebrate Proto-Oncogene, c- rel. Science 238: 692-694.
Steward R. (1989). Relocalization of the dorsal protein from the cytoplasm to the nucleus correlates with its function. Cell 59: 1179-1188.
Stock J. K., Giadrossi S., Casanova M., Brookes E., Vidal M., Koseki H., Brockdorff N., Fisher A. G., Pombo A. (2007). Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature Cell Biology 9: 1428-1435.
Sun Y., Nien C.Y., Chen K., Liu H.Y., Johnston J., Zeitlinger J., Rushlow C. (2015). Zelda overcomes the high intrinsic nucleosome barrier at enhancers during Drosophila zygotic genome activation. Genome Research 25: 1703-1714.
Suter D. M., Molina N., Gatfield D., Schneider K., Schibler U., Naef F. (2011). Mammalian Genes Are Transcribed with Widely Different Bursting Kinetics. Science 332: 472-474.
Tantale K., Garcia-Oliver E., Robert M.C., L’Hostis A., Yang Y., Tsanov N., Topno R., Gostan T., Kozulic-Pirher A., Basu-Shrivastava M., Mukherjee K., Slaninova V., Andrau J.C., Mueller F., Basyuk E., Radulescu O., Bertrand E. (2021). Stochastic pausing at latent HIV-1 promoters generates transcriptional bursting. Nature Communications 12: 4503.
The modENCODE Consortium Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F., Washietl S., Arshinoff B. I., Ay F., Meyer P. E., Robine N., Washington N. L., Di Stefano L., Berezikov E., Brown C. D., Candeias R., Carlson J. W., Carr A., Jungreis I., Marbach D., Sealfon R., Tolstorukov M. Y., Will S., Alekseyenko A. A., Artieri C., Booth B. W., Brooks A. N., Dai Q., Davis C. A., Duff M. O., Feng X., Gorchakov A. A., Gu T., Henikoff J. G., Kapranov P., Li R., MacAlpine H. K., Malone J., Minoda A., Nordman J., Okamura K., Perry M., Powell S. K., Riddle N. C., Sakai A., Samsonova A., Sandler J. E., Schwartz Y. B., Sher N., Spokony R., Sturgill D., van Baren M., Wan K. H., Yang L., Yu C., Feingold E., Good P., Guyer M., Lowdon R., Ahmad K., Andrews J., Berger B., Brenner S. E., Brent M. R., Cherbas L., Elgin S. C. R., Gingeras T. R., Grossman R., Hoskins R. A., Kaufman T. C., Kent W., Kuroda M. I., Orr-Weaver T., Perrimon N., Pirrotta V., Posakony J. W., Ren B., Russell S., Cherbas P., Graveley B. R., Lewis S., Micklem G., Oliver B., Park P. J., Celniker S. E., Henikoff S., Karpen G. H., Lai E. C., MacAlpine D. M., Stein L. D., White K. P., Kellis M., Acevedo D., Auburn R., Barber G., Bellen H. J., Bishop E. P., Bryson T. D., Chateigner A., Chen J., Clawson H., Comstock C. L. G., Contrino S., DeNapoli L. C., Ding Q., Dobin A., Domanus M. H., Drenkow J., Dudoit S., Dumais J., Eng T., Fagegaltier D., Gadel S. E., Ghosh S., Guillier F., Hanley D., Hannon G. J., Hansen K. D., Heinz E., Hinrichs A. S., Hirst M., Jha S., Jiang L., Jung Y. L., Kashevsky H., Kennedy C. D., Kephart E. T., Langton L., Lee O.K., Li S., Li Z., Lin W., Linder-Basso D., Lloyd P., Lyne R., Marchetti S. E., Marra M., Mattiuzzo N. R., McKay S., Meyer F., Miller D., Miller S. W., Moore R. A., Morrison C. A., Prinz J. A., Rooks M., Moore R., Rutherford K. M., Ruzanov P., Scheftner D. A., Senderowicz L., Shah P. K., Shanower G., Smith R., Stinson E. O., Suchy S., Tenney A. E., Tian F., Venken K. J. T., Wang H., White R., Wilkening J., Willingham A. T., Zaleski C., Zha Z., Zhang D., Zhao Y., Zieba J. (2010). Identification of Functional Elements and Regulatory Circuits by Drosophila modENCODE. Science 330: 1787-1797.
Thompson P. R., Wang D., Wang L., Fulco M., Pediconi N., Zhang D., An W., Ge Q., Roeder R. G., Wong J., Levrero M., Sartorelli V., Cotter R. J., Cole P. A. (2004). Regulation of the p300 HAT domain via a novel activation loop. Nature Structural & Molecular Biology 11: 308-315.
Thurman R. E., Rynes E., Humbert R., Vierstra J., Maurano M. T., Haugen E., Sheffield N. C., Stergachis A. B., Wang H., Vernot B., Garg K., John S., Sandstrom R., Bates D., Boatman L., Canfield T. K., Diegel M., Dunn D., Ebersol A. K., Frum T., Giste E., Johnson A. K., Johnson E. M., Kutyavin T., Lajoie B., Lee B.K., Lee K., London D., Lotakis D., Neph S., Neri F., Nguyen E. D., Qu H., Reynolds A. P., Roach V., Safi A., Sanchez M. E., Sanyal A., Shafer A., Simon J. M., Song L., Vong S., Weaver M., Yan Y., Zhang Z., Zhang Z., Lenhard B., Tewari M., Dorschner M. O., Hansen R. S., Navas P. A., Stamatoyannopoulos G., Iyer V. R., Lieb J. D., Sunyaev S. R., Akey J. M., Sabo P. J., Kaul R., Furey T. S., Dekker J., Crawford G. E., Stamatoyannopoulos J. A. (2012). The accessible chromatin landscape of the human genome. Nature 489: 75-82.
Tie F., Banerjee R., Fu C., Stratton C. A., Fang M., Harte P. J. (2016). Polycomb inhibits histone acetylation by CBP by binding directly to its catalytic domain. Proceedings of the National Academy of Sciences 113: E744-E753.
Tie F., Banerjee R., Stratton C. A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M. O., Scacheri P. C., Harte P. J. (2009). CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131-3141.
Trojanowski J., Frank L., Rademacher A., Mücke N., Grigaitis P., Rippe K. (2022). Transcription activation is enhanced by multivalent interactions independent of phase separation. Molecular Cell 82: 1878-1893.e10.
Tunnacliffe E., Chubb J. R. (2020). What Is a Transcriptional Burst?. Trends in Genetics 36: 288-297.
Ulianov S. V., Khrameeva E. E., Gavrilov A. A., Flyamer I. M., Kos P., Mikhaleva E. A., Penin A. A., Logacheva M. D., Imakaev M. V., Chertovich A., Gelfand M. S., Shevelyov Y. Y., Razin S. V. (2016). Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome Research 26: 70-84.
Vaid R., Wen J., Mannervik M. (2020). Release of promoter–proximal paused Pol II in response to histone deacetylase inhibition. Nucleic Acids Research 48: 4877-4890.
Vannam R., Sayilgan J., Ojeda S., Karakyriakou B., Hu E., Kreuzer J., Morris R., Herrera Lopez X. I., Rai S., Haas W., Lawrence M., Ott C. J. (2021). Targeted degradation of the enhancer lysine acetyltransferases CBP and p300. Cell Chemical Biology 28: 503-514.e12.
Vervoort S. J., Welsh S. A., Devlin J. R., Barbieri E., Knight D. A., Offley S., Bjelosevic S., Costacurta M., Todorovski I., Kearney C. J., Sandow J. J., Fan Z., Blyth B., McLeod V., Vissers J. H.A., Pavic K., Martin B. P., Gregory G., Demosthenous E., Zethoven M., Kong I. Y., Hawkins E. D., Hogg S. J., Kelly M. J., Newbold A., Simpson K. J., Kauko O., Harvey K. F., Ohlmeyer M., Westermarck J., Gray N., Gardini A., Johnstone R. W. (2021). The PP2A-Integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell 184: 3143-3162.e32.
Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., Afzal V., Ren B., Rubin E. M., Pennacchio L. A. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854-858.
Wagner E. J., Tong L., Adelman K. (2023). Integrator is a global promoter-proximal termination complex. Molecular Cell 83: 416-427.
Walters M. C., Fiering S., Eidemiller J., Magis W., Groudine M., Martin D. I. (1995). Enhancers increase the probability but not the level of gene expression.. Proceedings of the National Academy of Sciences 92: 7125-7129.
Wang H., Fan Z., Shliaha P. V., Miele M., Hendrickson R. C., Jiang X., Helin K. (2023). H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature 615: 339-348.
Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T.Y., Peng W., Zhang M. Q., Zhao K. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genetics 40: 897-903.
Weinert B. T., Narita T., Satpathy S., Srinivasan B., Hansen B. K., Schölz C., Hamilton W. B., Zucconi B. E., Wang W. W., Liu W. R., Brickman J. M., Kesicki E. A., Lai A., Bromberg K. D., Cole P. A., Choudhary C. (2018). Time-Resolved Analysis Reveals Rapid Dynamics and Broad Scope of the CBP/p300 Acetylome. Cell 174: 231-244.e12.
Wong E. S., Zheng D., Tan S. Z., Bower N. I., Garside V., Vanwalleghem G., Gaiti F., Scott E., Hogan B. M., Kikuchi K., McGlinn E., Francois M., Degnan B. M. (2020). Deep conservation of the enhancer regulatory code in animals. Science 370: eaax8137.
Xi H., Shulha H. P., Lin J. M., Vales T. R., Fu Y., Bodine D. M., McKay R. D. G., Chenoweth J. G., Tesar P. J., Furey T. S., Ren B., Weng Z., Crawford G. E. (2007). Identification and Characterization of Cell Type–Specific and Ubiquitous Chromatin Regulatory Structures in the Human Genome. PLoS Genetics 3: e136.
Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H. (2006). P-TEFb-Mediated Phosphorylation of hSpt5 C-Terminal Repeats Is Critical for Processive Transcription Elongation. Molecular Cell 21: 227-237.
Yang Z., Yik J. H.N., Chen R., He N., Jang M. K., Ozato K., Zhou Q. (2005). Recruitment of P-TEFb for Stimulation of Transcriptional Elongation by the Bromodomain Protein Brd4. Molecular Cell 19: 535-545.
Yokoshi M., Segawa K., Fukaya T. (2020). Visualizing the Role of Boundary Elements in Enhancer-Promoter Communication. Molecular Cell 78: 224-235.e5.
Zaugg J. B., Sahlén P., Andersson R., Alberich-Jorda M., de Laat W., Deplancke B., Ferrer J., Mandrup S., Natoli G., Plewczynski D., Rada-Iglesias A., Spicuglia S. (2022). Current challenges in understanding the role of enhancers in disease. Nature Structural & Molecular Biology 29: 1148-1158.
Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R. A. (2007a). RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature Genetics 39: 1512-1516.
Zeitlinger J., Zinzen R. P., Stark A., Kellis M., Zhang H., Young R. A., Levine M. (2007b). Whole-genome ChIP–chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes & Development 21: 385-390.
Zhang T., Zhang Z., Dong Q., Xiong J., Zhu B. (2020). Histone H3K27 acetylation is dispensable for enhancer activity in mouse embryonic stem cells. Genome Biology 21: 45.
Zhang Y., Brown K., Yu Y., Ibrahim Z., Zandian M., Xuan H., Ingersoll S., Lee T., Ebmeier C. C., Liu J., Panne D., Shi X., Ren X., Kutateladze T. G. (2021). Nuclear condensates of p300 formed though the structured catalytic core can act as a storage pool of p300 with reduced HAT activity. Nature Communications 12: 4618.
Zoller B., Little S. C., Gregor T. (2018). Diverse Spatial Expression Patterns Emerge from Unified Kinetics of Transcriptional Bursting. Cell 175: 835-847.e25.