Int. J. Dev. Biol. 67: 147 - 153 (2023)
The stem cell transcription factor ZFP296 transforms NIH3T3 cells and promotes anchorage-independent growth of cancer cells
Open Access | Original Article | Published: 30 January 2024
Abstract
Cancer cells and embryonic stem (ES) cells share several biological properties, suggesting that some genes expressed in ES cells may play an important role in cancer cell growth. In this study, we investigated the possible role of zinc finger protein 296 (ZFP296), a transcription factor expressed in ES cells, in cancer development. First, we found that overexpression of Zfp296 in NIH3T3 mouse fibroblasts induced two phenomena indicative of cell transformation: enhanced proliferation under low-serum conditions and anchorage-independent growth. We also found that Zfp296 expression was upregulated in the tumor area of a mouse model of colon carcinogenesis. In addition, the expression levels of ZFP296 in various human cell lines were generally low in normal cells and relatively high in cancer cells. Finally, using a soft agar assay, we found that overexpression of ZFP296 promoted the anchorage-independent growth of cancer cells, while its knockdown had the opposite effect. Overall, these results suggest a possible role of the ES-specific transcription factor ZFP296 in cancer.
Keywords
ZNF296, anchorage-independent growth, oncogene, embryonic stem cells, tumor
Introduction
Embryonic stem (ES) cells are established from the inner cell mass of early mammalian embryos and can undergo self-renewal in vitro while maintaining multipotency. Notably, ES cells share many biological characteristics with cancer cells (Ben-David and Benvenisty, 2011; Koide, 2014), such as an enhanced proliferation rate and high telomerase activity, which allows them to proliferate indefinitely. Furthermore, when injected into nude mice, ES cells can give rise to benign tumors called teratomas. In addition, several signaling pathways, including the STAT3 and Wnt pathways, play important roles in both ES cell self-renewal and cancer cell growth (Dreesen and Brivanlou 2007; Koide, 2014). These similarities suggest that some molecules involved in the self-renewal of ES cells may also play important roles in cancer cell proliferation. In our previous work, we searched for novel oncogenes in self-renewing ES cells and reported the identification of zinc finger protein 57 (ZFP57), a transcription factor that transforms normal cells and promotes the tumorigenic and metastatic potential of cancer cells (Tada et al., 2015; Shoji et al., 2019).
Here, we focused on another ES cell-expressed transcription factor, ZFP296 (also known as ZNF296 or ZNF342), which belongs to the same class of transcription factors as ZFP57, and investigated its possible involvement in cancer development. We found that ZFP296 was expressed at relatively high levels in human cancer cell lines. Furthermore, overexpression of ZFP296 transformed normal mouse fibroblasts and promoted the anchorage-independent growth of human cancer cells, whereas its suppression inhibited such growth. These results suggest that ZFP296 plays a role in cell transformation and tumor growth.
Results
Zfp296 is expressed at a high level in self-renewing mouse ES cells
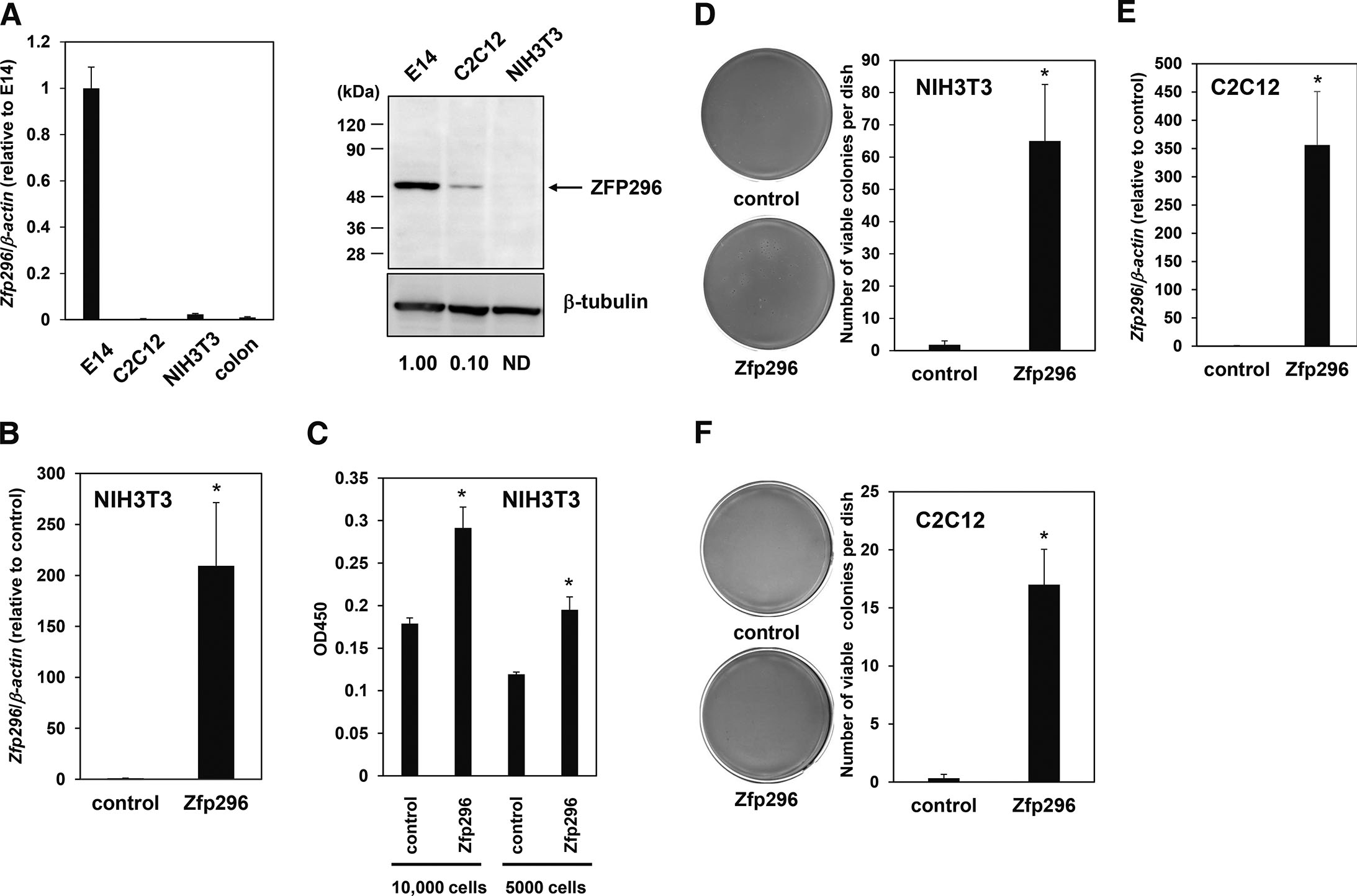
We reported previously that Zfp296 is expressed in self-renewing mouse ES cells and its expression level is reduced upon differentiation (Fujii et al., 2013). Similarly, self-renewal-specific expression of ZFP296 has been reported in human ES cells (Fischedick et al., 2012). Here, real-time RT-qPCR analyses revealed that the expression level of Zfp296 mRNA in self-renewing mouse ES cells (E14) was substantially higher than that found in mouse fibroblasts (NIH3T3 cells), myoblasts (C2C12 cells), and steady-state colon tissues (Fig. 1A, left panel). Similarly, the expression level of ZFP296 protein in E14 cells was much higher than in NIH3T3 and C2C12 cells (Fig. 1A, right panel).
Fig. 1. Zfp296 is expressed at a high level in self-renewing mouse ES cells and transforms mouse fibroblasts in vitro.
(A) Real-time RT-qPCR (left panel) and Western blot (right panel) analyses comparing the expression level of Zfp296 in self-renewing ES cells (E14) with those in C2C12 cells (mouse myoblast cell line), NIH3T3 cells (mouse fibroblast cell line), and mouse colon tissue. Data were normalized to the expression level in E14 cells. The numbers below the gel indicate the expression level of ZFP296 in each cell line relative to that in E14, which was calculated by dividing the density of ZFP296 bands by that of β-tubulin bands, followed by setting the value of E14 as 1. ND, not detected. (B) Real-time RT-qPCR analyses confirming the overexpression of Zfp296 in NIH3T3 cells. Data were normalized to the level in control cells. (C) WST-1 assay to evaluate the effect of overexpression of Zfp296 on the growth of NIH3T3 cells under low-serum conditions. The indicated numbers of cells were seeded into 96-well plates and cultured for 3 days. Viable cells were measured using a commercial WST-1 assay kit. (D) Soft agar assay to evaluate the effect of overexpression of Zfp296 on anchorage-independent growth of NIH3T3 cells. The cells (6,000) were cultured in soft agar, and viable colonies were stained with MTT and counted. Representative scanned images of NIH3T3 cell soft agar plates stained with MTT (left panel) and the number of stained colonies counted using the Image-J software (right panel) are shown. (E) Real-time RT-qPCR analyses confirming the overexpression of Zfp296 in C2C12 cells. Data were normalized to the level in control cells. (F) Soft agar assay to evaluate the effect of overexpression of Zfp296 on anchorage-independent growth of C2C12 cells. The cells (12,000) were cultured in soft agar and viable colonies were stained with MTT and counted. Representative scanned images (left panel) and the number of stained colonies (right panel) are shown. In all experiments, data are represented as the mean ± standard error (n ≥ 3); *P < 0.05.
ZFP296 transforms normal mouse cells
NIH3T3 is a normal fibroblast cell line isolated from fetal mouse skin. To determine whether ZFP296 can transform normal cells into cancer cells, we overexpressed Zfp296 in NIH3T3 cells (Fig. 1B and S1A) and examined the acquisition of two characteristics of cancer cells, growth under low-serum conditions and anchorage-independent growth. Overexpression of Zfp296 promoted the growth of NIH3T3 cells in low-serum (0.5% calf serum) medium (Fig. 1C). NIH3T3 cells can hardly proliferate in soft agar, but overexpression of Zfp296 promoted their ability to grow in an anchorage-independent manner (Fig. 1D). Overexpression of Zfp296 also induced anchorage-independent growth of another normal cell line, C2C12 myoblast (Fig. 1 E,FY and S1B). These results suggest that ZFP296 can transform normal cells into cancer cells.
Zfp296 is upregulated in a mouse tumor model
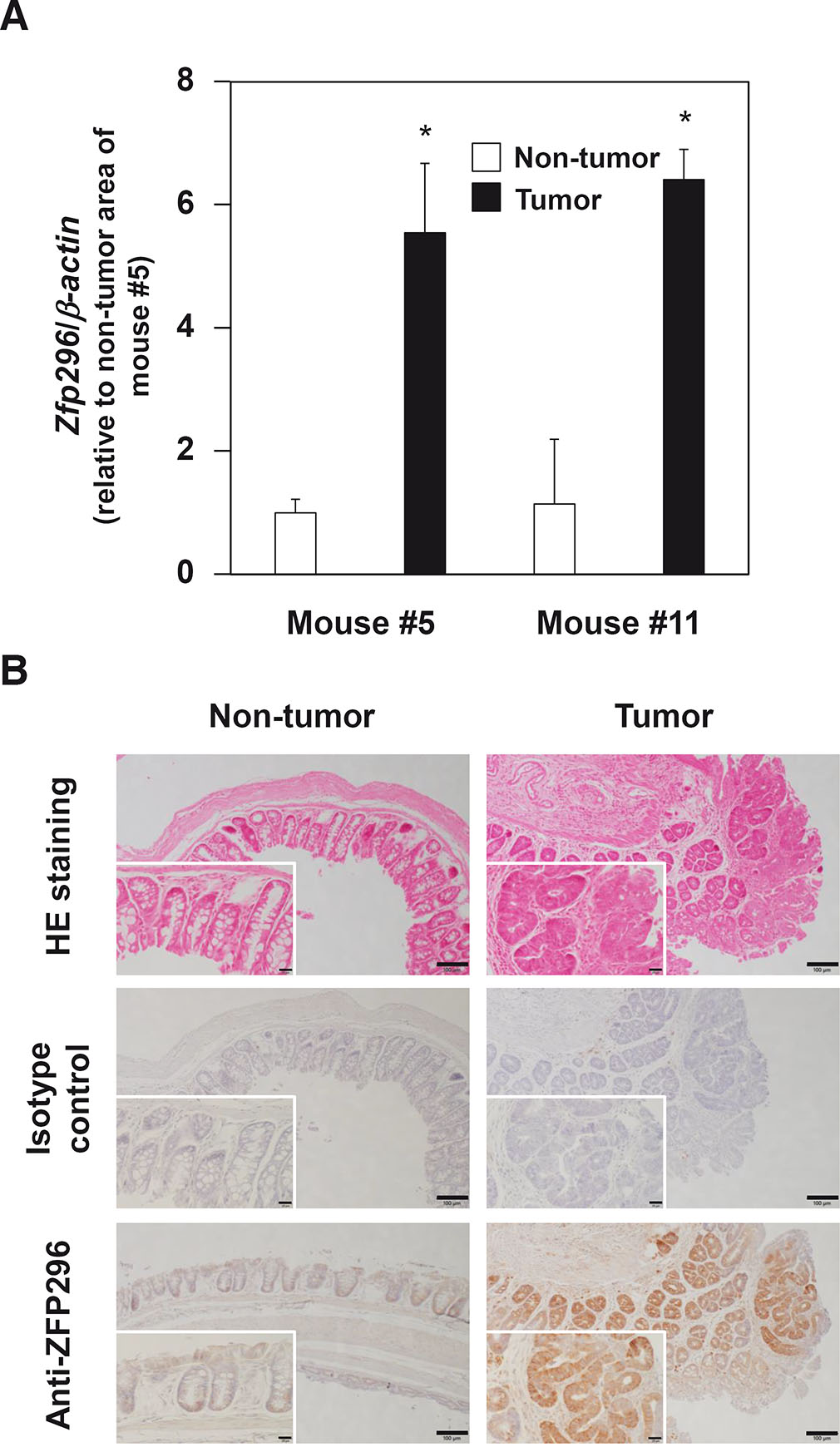
To explore its possible involvement in cancer development further, the expression level of Zfp296 was examined in a mouse colon cancer model in which colitis-associated colorectal cancer was artificially induced by exposure to a single hit of the carcinogen AOM, followed by repeated exposures to the inflammatory agent DSS (De Robertis et al., 2011). Real-time RT-qPCR and immunohistochemistry analysis revealed that the expression level of Zfp296 was higher in tumor areas than in non-tumor areas (Fig. 2), suggesting the involvement of Zfp296 in in vivo cancer development in mice.
Fig. 2. Zfp296 is expressed at a high level in the tumor areas of AOM/DSS-treated mice.
(A) Real-time RT-qPCR analysis was performed on each sample in triplicate. Data are represented as the mean ± standard error. The value for the non-tumor area of mouse #5 was set to 1. *P < 0.05. (B) Representative images of H&E and immunoreactive ZFP296 staining in murine colon sections are shown. Differences in ZFP296 expression level are found between tumor and non-tumor areas (scale bar, 100 µm). Inserts show magnified images (Scale bar, 20 µm).
ZFP296 is expressed highly in cancer cell lines
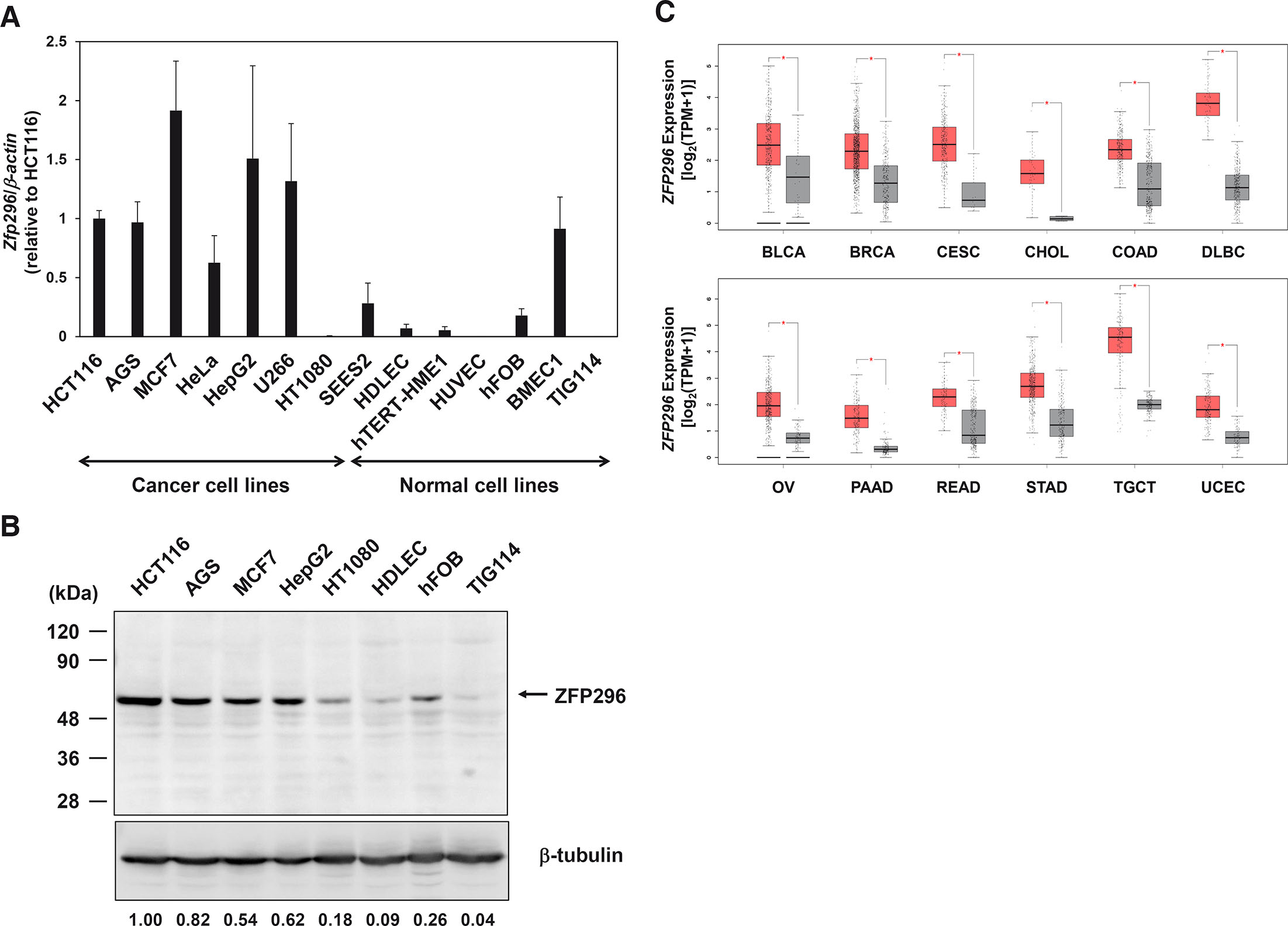
Next, we examined the expression levels of ZFP296 in various human cancer and non-cancer cell lines (Fig. 3A). ZFP296 mRNA was expressed at a high level in six of the seven cancer cell lines examined, although it was expressed at a low level in HT1080 fibrosarcoma cells. By contrast, with the exception of BMEC1 (bone marrow microvascular endothelium) cells, ZFP296 mRNA was expressed at a relatively low level in non-cancer cell lines. The expression level of ZFP296 mRNA in human ES cells was generally lower than that found in cancer cells but higher than that expressed in normal cells. Similarly, ZFP296 protein was highly expressed in cancer cell lines except HT1080, but low in non-cancer cell lines (Fig. 3B). Furthermore, based on data from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx), we compared the expression levels of this transcription factor in cancer and normal tissues using GEPIA2 software (Tang et al., 2019), and found that ZFP296 mRNA was upregulated in several cancers, including bladder, breast, colon, ovarian, pancreatic and stomach cancers (Fig. 3C and S2). Overall, these results suggest that ZFP296 may be involved in tumorigenesis.
Fig. 3. Expression levels of ZFP296 in various human cancer and non-cancer cell lines.
(A) Real-time RT-qPCR analyses were carried out using total RNA extracted from the following cell lines: HCT116 (colon cancer), AGS (gastric cancer), MCF7 (breast cancer), HeLa (cervical cancer), HepG2 (liver cancer), U266 (myeloma), HT1080 (fibrosarcoma), SEES2 (ES cells), HDLEC (dermal lymphatic endothelium), hTERT-HME1 (mammary epithelium), HUVEC (umbilical vein endothelium), hFOB (fetal osteoblast), BMEC1 (bone marrow microvascular endothelium), and TIG114 (fibroblast). Data were normalized to the expression level in HCT116 cells and are represented as the mean ± standard error (n ≥ 3). (B) Western blot analyses were carried out using total cell lysate from the indicated cell lines. The numbers below the gel indicate the expression level of ZFP296 in each cell line relative to that in HCT116. (C) Expression of ZFP296 mRNA in several human tumors (red box) and normal tissues (grey box). All data are shown in Fig. S2, and only cancers with significantly higher expression of Zfp296 in tumor tissue compared to normal tissue are shown here. Abbreviations: BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, diffuse large B-cell lymphoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; READ, rectum adenocarcinoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; UCEC, uterine corpus endometrial carcinoma. *P<0.01.
ZFP296 is involved in the anchorage-independent growth of cancer cells
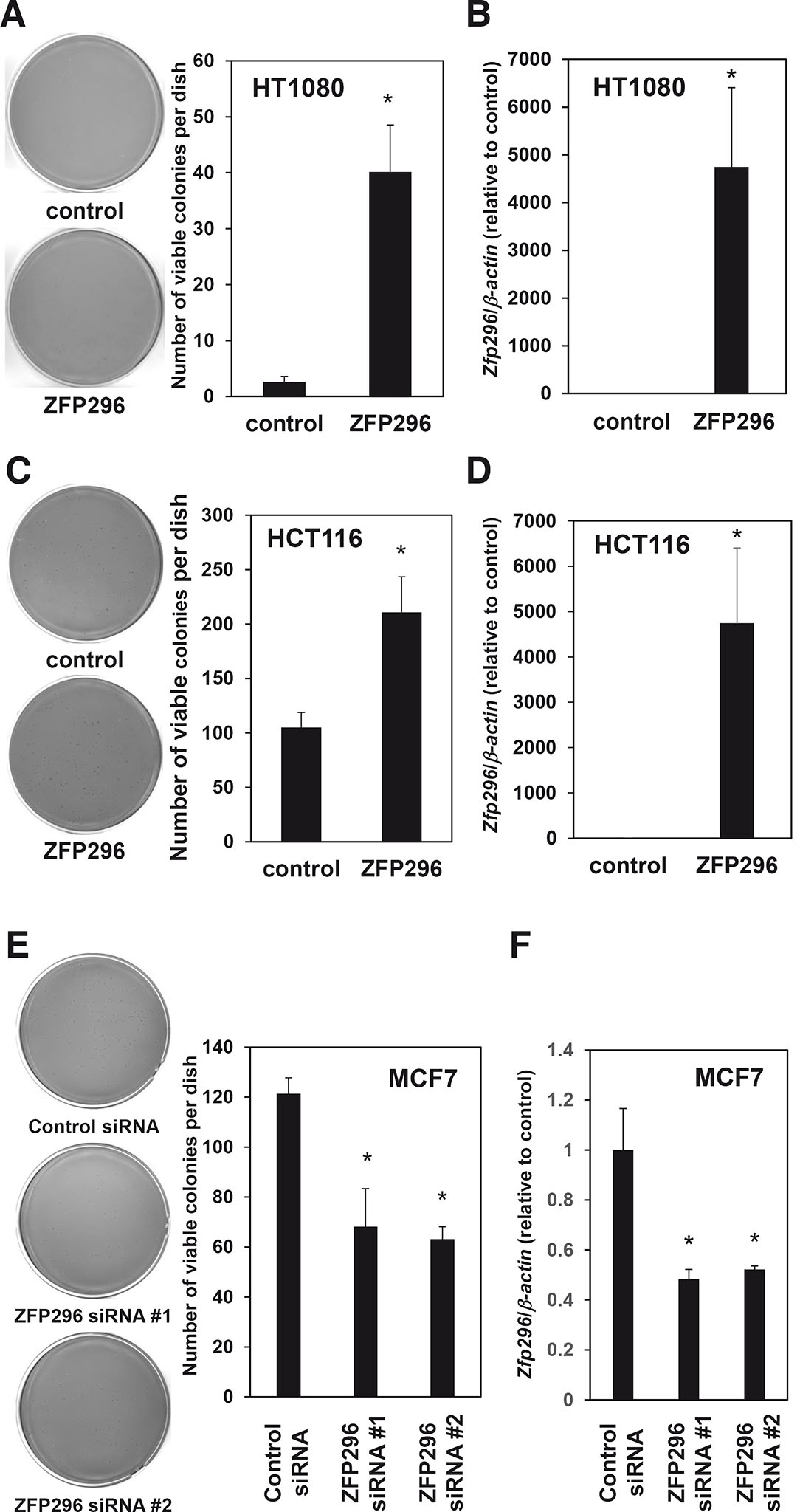
To examine its involvement in the tumor progression of cancer cells, we examined the effect of overexpression of ZFP296 on anchorage-independent growth of the malignant HT1080 fibrosarcoma cell line, which had relatively low levels of ZFP296 expression (Fig. 3 A,B). Overexpression of ZFP296 enhanced the anchorage-independent proliferative ability of HT1080 cells significantly (Fig. 4 A,B, and S1C). Similarly, overexpression of this transcription factor increased the anchorage-independent growth of HCT116 cells (Fig. 4 C,D and S1D). These data suggest that ZFP296 can promote anchorage-independent growth of cancer cells and contribute to tumor progression.
Fig. 4. ZFP296 is involved in the anchorage-independent growth of cancer cells.
(A) The effect of overexpression of ZFP296 on the anchorage-independent growth of HT1080 cells. Cells (12,000) were cultured in soft agar and the numbers of colonies stained with MTT were counted. Representative scanned images (left panel) and the number of stained colonies (right panel) are shown. (B) Real-time RT-qPCR analysis confirming the overexpression of ZFP296 in HT1080 cells. Data were normalized to the level in control cells. (C) The effect of overexpression of ZFP296 on the anchorage-independent growth of HCT116 cells. Cells (1,200) were cultured in soft agar, and the number of colonies stained with MTT was counted. Representative scanned images (left panel) and the number of stained colonies (right panel) are shown. (D) Real-time RT-qPCR analysis confirming the overexpression of ZFP296 in HCT116 cells. Data were normalized to the level in control cells. (E) The effect of siRNA-mediated knockdown of ZFP296 on anchorage-independent growth of MCF7 cells. Cells (600) were cultured in soft agar and the numbers of colonies stained with MTT were counted. Representative scanned images (left panel) and the number of stained colonies (right panel) are shown. (F) Real-time RT-qPCR analysis confirming the knockdown of ZFP296 in MCF7 cells. Data were normalized to the level in control cells. In all experiments, data are represented as the mean ± standard error (n = 3); *P < 0.05.
Next, to determine whether ZFP296 is required for the anchorage-independent growth of cancer cells, we knocked down ZFP296 expression in MCF7 cells, which had high levels of the transcription factor (Fig. 3). Suppression of ZFP296 using two different siRNAs inhibited the anchorage-independent growth of MCF7 cells significantly (Fig. 4 E,F and S1E). Taken together, these results suggest that ZFP296 is involved in the tumorigenesis of cancer cells.
Discussion
In this study, we examined the possible involvement of ZFP296 in cancer. We found that overexpression of Zfp296 transformed mouse normal fibroblasts and myoblasts. In addition, the use of a mouse model of colorectal cancer revealed higher expression of Zfp296 in tumor areas than in non-tumor areas. In human cell lines, ZFP296 expression was higher for cancer cells than for normal cells. These results suggest that ZFP296 plays a role in tumorigenesis. In support of this hypothesis, we also found that overexpression of ZFP296 promoted the anchorage-independent growth of the human fibrosarcoma-derived cell line HT1080 and colon cancer-derived cell line HCT116. Furthermore, knockdown of ZFP296 expression inhibited the anchorage-independent growth of breast cancer-derived MCF7 cells.
Several research groups have reported various biological functions of ZFP296, each from their own perspective. One group reported that ZFP296 is more abundant in patients with latent tuberculosis than with tuberculosis (Gliddon et al., 2021). Other groups have reported that ZFP296 plays a role in germline specification by modulating Wnt activity (Hackett et al., 2018), negatively regulates global H3K9 methylation in embryonic development as a component of heterochromatin (Matsuura et al., 2017), and interacts with the nucleosome remodeling and deacetylase (NuRD) complex to regulates genome-wide NuRD localization (Kloet et al., 2018). ZFP296 is also upregulated in response to increased extracellular zinc (Zaman et al., 2021).
Regarding its relationship to cancer, our present results suggest that ZFP296 is involved in the anchorage-independent growth of fibrosarcoma, colon, and breast cancer cells. Similarly, high levels of ZFP296 expression in acute lymphoblastic leukemia are associated with a poor outcome (Poland et al., 2009). On the other hand, the ZFP296 gene is highly methylated in astrocytic and oligodendroglial tumors, and ZFP296 expression is decreased in primary oligodendrogliomas (Hong et al., 2003; Zheng et al., 2011). Therefore, the involvement of ZFP296 seems to vary depending on the type of cancer. Notably, ZFP296 can promote the formation of induced pluripotent cells (Fischedick et al., 2012), which may be related to the cell growth-promoting activity of ZFP296 described in our current study.
Although we found here that ZFP296 can promote anchorage-independent growth, the molecular mechanism underlying this effect is unknown. We reported previously that ZFP296 binds to KLF4 and suppresses its activity (Fujii et al., 2013). KLF4 acts as a tumor suppressor gene in various cancers (Taracha-Wisniewska et al., 2020). For example, in one study, forced expression of KLF4 in a hepatocellular cancer cell line reduced anchorage-independent growth in soft agar (Lin et al., 2012), while another study found that overexpression of KLF4 suppressed the tumor formation of cervical carcinoma cells in nude mice (Yang and Zheng, 2012). KLF4 expression is reduced in colorectal cancer, and KLF4 deficiency is an independent predictor of survival and disease recurrence (Patel et al., 2010). Therefore, ZFP296 might promote the anchorage-independent growth of certain types of cancer cells by suppressing KLF4.
Preliminary results from our lab have suggested that ZFP296 upregulates ZFP57, albeit slightly, when overexpressed in NIH3T3 cells (data not shown). Since we have demonstrated previously that overexpression of ZFP57 in NIH3T3 cells stimulates their anchorage-independent growth (Tada et al., 2015), ZFP296 may transform NIH3T3 cells via the upregulation of ZFP57. In addition, ZFP296 negatively regulates global H3K9 methylation (Matsuura et al., 2017), suggesting that it may also promote growth via epigenetic regulation. We are currently analyzing the molecular mechanism of ZFP296-induced cellular oncogenesis and promotion of cancer cell proliferation.
Materials and methods
Cell culture
NIH3T3 cells were obtained from the RIKEN Cell Bank (Wako, Japan) and were cultured in Dulbecco’s modified Eagle medium (DMEM; Nacalai Tesque, Kyoto, Japan) containing 10% calf serum. HT1080 cells obtained from the Japanese Collection of Research Bioresources (JCRB, Osaka, Japan) and MCF7 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). The mouse ES cell line E14TG2a was obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured as described previously (Tada et al., 2015). The human ES cell line SEES2 (Akutsu et al., 2015) was maintained on irradiated mouse embryonic fibroblast feeder layers in DMEM containing 20% knockout serum replacement, 0.1 mM non-essential amino acids, 2 mM GlutaMAX-I, 50 U/mL penicillin, 50 μg/mL streptomycin, and 8 ng/mL recombinant human bFGF (all from Life Technologies, Carlsbad, CA, USA). All cells were cultured in a humidified atmosphere with 5% CO2 at 37°C. Culture of other cell lines is described in the Supplemental Information.
Overexpression and knockdown of ZFP296
The mammalian expression vector pCAGIPj2 was constructed by replacing the CMV promoter region of pIRESpuro3 (Takara Bio, Shiga, Japan) with the CAG promoter region (Niwa et al., 1991) of pEBMulti-Puro (FUJIFILM Wako Chemicals, Osaka, Japan) and modifying the multiple cloning site sequence. For overexpression in cell lines, the coding region of myc-tagged mouse Zfp296 was inserted into pCAGIP (Tada et al., 2015), and that of myc-tagged human ZFP296 was inserted into pCAGIPj2 to obtain pCAGIP-mycZfp296 and pCAGIPj2-mycZFP296, respectively. The mouse and human expression vectors were introduced into NIH3T3, C2C12, HCT116, and HT1080 cells, respectively, using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA), and were selected with 0.8, 1.5, 0.3 and 0.25 µg/mL puromycin, respectively.
For knockdown experiments, MISSION siRNAs targeting ZFP296, or a universal negative control (Sigma-Aldrich, St. Louis, MO, USA), were transfected into MCF7 cells using Lipofectamine 2000.
Establishment of the AOM/DSS model of colon carcinogenesis
Eight to 12-weeks-old male C57BL6 mice were purchased from Japan SLC Inc. (Hamamatsu, Japan) and housed under specific pathogen-free conditions. To establish the azoxymethane/dextran sulfate sodium (AOM/DSS) model of colon carcinogenesis, the C57BL/6 mice were injected intraperitoneally with 12.5 mg/kg AOM (FUJIFILM Wako Chemicals) dissolved in physiological saline. Seven days later, the mice were given drinking water containing 2% DSS (molecular weight =36,000–50,000 Da; ICN Biomedicals, Costa Mesa, CA, USA) for 5 days, followed by regular water for 16 days. This cycle (5 days of DSS followed by 16 days of regular water) was repeated three times, and mice were sacrificed on day 77. Colons were removed and flushed with phosphate-buffered saline. The institutional Animal Care and Use Committee of Juntendo University School of Medicine, Tokyo, Japan, approved the animal procedure protocols. All animal experiments complied with the ARRIVE guidelines and carried out in accordance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Real-time RT-qPCR
Total RNA was prepared using a Total RNA Purification Kit (BioElegen Technology, Taichung, Taiwan) and subjected to reverse transcription using ReverTra Ace (Toyobo, Osaka, Japan). Real-time RT-qPCR was performed using a 7500 Fast or QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific). Primer sets used for PCR are shown in Table S1. In all experiments, β-actin was used as an internal control, and the expression levels of each gene were calculated by ΔΔCt method.
Western blot analysis
Cell lysates were subjected to SDS–polyacrylamide gel electrophoresis and electrophoretically transferred to nitrocellulose membranes (Pall, Port Washington, NY, USA). The membranes were then probed with either anti-ZNF296 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-β-tubulin antibody (FUJIFILM Wako Chemicals), and signals were visualized using ECL Western blotting detection reagents (Cytiva, Marlborough, MA, USA) with the LAS-4000 image analyzer (GE Health Care, Piscataway, NJ, USA). In all experiments, densitometric quantification of band intensity was performed using Image-J software (NIH, Bethesda, MD, USA) and normalized relative to the band intensity of β-tubulin.
Immunohistochemistry
Colon tissues were fixed in 4% paraformaldehyde, paraffin blocks were generated, and 4 µm thick sections were cut. Immunohistochemical staining was performed using Mouse on Mouse (M.O.M.) Immunodetection Kit (Vector Laboratories, Newark, CA, USA). Following the antigen retrieval using Tris-EDTA buffer (pH 9.0) at 121o C, endogenous peroxidase activity was blocked using 0.3% H2O2 in methanol for 30 min. Avidin/Biotin blocking was performed using Avidin/Biotin Blocking Kit (Vector Laboratories). Tissue sections were blocked with M.O.M. Mouse IgG Blocking Reagent and stained with anti-ZNF296 antibody (Santa Cruz Biotechnology) for 30 min at room temperature. The sections were then stained with M.O.M. Biotinylated Anti-Mouse IgG reagent. Tissues were incubated with Streptavidin/HRP (DAKO, Denmark) for 30 min at room temperature. Signals were developed by reaction with 3,3′-diaminobenzidine, tetrahydrochloride (DAB; Dojindo, Kumamoto, Japan). Tissue sections were counterstained with hematoxylin and eosin (H&E). Images were taken using an Olympus BX53 light microscope.
WST-1 assay
Cell viability was examined using the WST-1 Cell Proliferation Assay Kit (Takara Bio), according to the manufacturer’s protocol. Briefly, cells were seeded into 96-well plates at a density of 5000 or 10,000 cells/well and were cultured in DMEM containing 0.5% CS for 3 days. Subsequently, WST-1 was added to each well and absorbance at 450 nm was measured using a SpectraMax 340PC384 microplate reader (Molecular Devices, Sunnyvale, CA, USA).
Soft agar assay
For the soft agar assay, cells were suspended in 2.4 mL of culture medium containing 0.5% SeaPlaque agarose (Lonza, Rockland, ME, USA) and then overlaid on 3 mL of culture medium containing 0.53% SeaPlaque agarose in 6 cm petri dishes. Cultures were maintained for 1 to 2 weeks. Viable colonies were stained with 3-(4,5-di-methylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Nacalai Tesque) and counted using Image-J software.
Statistical analysis
All statistical analyses were performed by a two-tailed Student's t test, and values of P < 0.05 were defined as statistically significant unless otherwise stated. In the figures, bars represent means, and error bars represent standard errors (SE) calculated from numerical data.
Supplementary Material
Acknowledgements
This work was supported in part by JSPS KAKENHI Grant Numbers JP19K07672, JP22F21773, JP21K08404, JP22KF0337, Grants from The Japanese Society of Hematology Research Grant (KH), the society of iodine science (KH), Radiation effects association (KH), Heiwa Nakajima Foundation (BH), Institute for Environmental and Gender-Specific Medicine (BH), and International Joint Usage/Research Center, the Institute of Medical Science, the University of Tokyo (KH, BH). We thank the Laboratory of Cell Biology and the Laboratory of Morphology and Image Analysis, Biomedical Research Core Facilities, Juntendo University Graduate School of Medicine, for technical assistance.
Abbreviations
AOM, azoxymethane ; CS, calf serum ; DMEM, Dulbecco’s modified Eagle medium ; DSS, dextran sulfate sodium ; ES, embryonic stem ; FBS, fetal bovine serum ; iPS, induced pluripotent stem ; NuRD, nucleosome remodeling and deacetylase ; ZFP, zinc finger protein ;References
Akutsu H., Machida M., Kanzaki S., Sugawara T., Ohkura T., Nakamura N., Yamazaki-Inoue M., Miura T., Vemuri M. C., Rao M. S., Miyado K., Umezawa A. (2015). Xenogeneic-free defined conditions for derivation and expansion of human embryonic stem cells with mesenchymal stem cells. Regenerative Therapy 1: 18-29.
Ben-David U., Benvenisty N. (2011). The tumorigenicity of human embryonic and induced pluripotent stem cells. Nature Reviews Cancer 11: 268-277.
De Robertis M., Massi E., Poeta M. L., Carotti S., Morini S., Cecchetelli L., Signori E., Fazio V. M., (2011). The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J. Carcinog. 10: 9.
Dreesen O., Brivanlou A. H. (2007). Signaling Pathways in Cancer and Embryonic Stem Cells. Stem Cell Reviews 3: 7-17.
Fischedick G., Klein D. C., Wu G., Esch D., Höing S., Han D. W., Reinhardt P., Hergarten K., Tapia N., Schöler H. R., Sterneckert J. L. (2012). Zfp296 Is a Novel, Pluripotent-Specific Reprogramming Factor. PLoS ONE 7: e34645.
Fujii Y., Kakegawa M., Koide H., Akagi T., Yokota T. (2013). Zfp296 is a novel Klf4-interacting protein and functions as a negative regulator. Biochemical and Biophysical Research Communications 441: 411-417.
Gliddon H. D., Kaforou M., Alikian M., Habgood-Coote D., Zhou C., Oni T., Anderson S. T., Brent A. J., Crampin A. C., Eley B., Heyderman R., Kern F., Langford P. R., Ottenhoff T. H. M., Hibberd M. L., French N., Wright V. J., Dockrell H. M., Coin L. J., Wilkinson R. J., Levin M. (2021). Identification of Reduced Host Transcriptomic Signatures for Tuberculosis Disease and Digital PCR-Based Validation and Quantification. Frontiers in Immunology 12: 637164.
Hackett J. A., Huang Y., Günesdogan U., Gretarsson K. A., Kobayashi T., Surani M. A. (2018). Tracing the transitions from pluripotency to germ cell fate with CRISPR screening. Nature Communications 9: 4292.
Hong C., Bollen A. W., Costello J. F., (2003). The contribution of genetic and epigenetic mechanisms to gene silencing in oligodendrogliomas. Cancer Research 63: 7600-7605.
Kloet S. L., Karemaker I. D., van Voorthuijsen L., Lindeboom R. G. H., Baltissen M. P., Edupuganti R. R., Poramba-Liyanage D. W., Jansen P. W. T. C., Vermeulen M. (2018). NuRD-interacting protein ZFP296 regulates genome-wide NuRD localization and differentiation of mouse embryonic stem cells. Nature Communications 9: 4588.
Koide H., (2014). Embryonic stem cells and oncogenes. In Pluripotent stem cell biology - advances in mechanisms, methods and models. (Ed. Atwood C. S., Meethal S. V., ) InTech, Rijeka.
Lin Z.S., Chu H.C., Yen Y.C., Lewis B. C., Chen Y.W. (2012). Krüppel-Like Factor 4, a Tumor Suppressor in Hepatocellular Carcinoma Cells Reverts Epithelial Mesenchymal Transition by Suppressing Slug Expression. PLoS ONE 7: e43593.
Matsuura T., Miyazaki S., Miyazaki T., Tashiro F., Miyazaki J. (2017). Zfp296 negatively regulates H3K9 methylation in embryonic development as a component of heterochromatin. Scientific Reports 7: 12462.
Niwa H., Yamamura K., Miyazaki J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193-199.
Patel N. V., Ghaleb A. M., Nandan M. O., Yang V. W. (2010). Expression of the Tumor Suppressor Krüppel-Like Factor 4 as a Prognostic Predictor for Colon Cancer. Cancer Epidemiology, Biomarkers & Prevention 19: 2631-2638.
Poland K. S., Shardy D. L., Azim M., Naeem R., Krance R. A., Dreyer Z.A. E., Neeley E. S., Zhang N., Qiu Y. H., Kornblau S. M., Plon S. E. (2009). Overexpression of ZNF342 by juxtaposition with MPO promoter/enhancer in the novel translocation t(17;19)(q23;q13.32) in pediatric acute myeloid leukemia and analysis of ZNF342 expression in leukemia . Genes, Chromosomes and Cancer 48: 480-489.
Shoji Y., Takamura H., Ninomiya I., Fushida S., Tada Y., Yokota T., Ohta T., Koide H. (2019). The Embryonic Stem Cell-Specific Transcription Factor ZFP57 Promotes Liver Metastasis of Colorectal Cancer. Journal of Surgical Research 237: 22-29.
Tada Y., Yamaguchi Y., Kinjo T., Song X., Akagi T., Takamura H., Ohta T., Yokota T., Koide H. (2015). The stem cell transcription factor ZFP57 induces IGF2 expression to promote anchorage-independent growth in cancer cells. Oncogene 34: 752-760.
Tang Z., Kang B., Li C., Chen T., Zhang Z. (2019). GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Research 47: W556-W560.
Taracha-Wisniewska A., Kotarba G., Dworkin S., Wilanowski T. (2020). Recent Discoveries on the Involvement of Krüppel-Like Factor 4 in the Most Common Cancer Types. International Journal of Molecular Sciences 21: 8843.
Yang W.T., Zheng P.S. (2012). Krüppel‐like factor 4 functions as a tumor suppressor in cervical carcinoma. Cancer 118: 3691-3702.
Zaman M. S., Barman S. K., Corley S. M., Wilkins M. R., Malladi C. S., Wu M. J. (2021). Transcriptomic insights into the zinc homeostasis of MCF-7 breast cancer cells via next-generation RNA sequencing. Metallomics 13: mfab026.
Zheng S., Houseman E. A., Morrison Z., Wrensch M. R., Patoka J. S., Ramos C., Haas-Kogan D. A., McBride S., Marsit C. J., Christensen B. C., Nelson H. H., Stokoe D., Wiemels J. L., Chang S. M., Prados M. D., Tihan T., Vandenberg S. R., Kelsey K. T., Berger M. S., Wiencke J. K. (2011). DNA hypermethylation profiles associated with glioma subtypes and EZH2 and IGFBP2 mRNA expression. Neuro-Oncology 13: 280-289.