Int. J. Dev. Biol. 68: 211 - 222 (2024)
Special Issue: Developmental Biology in Nordic Countries
Fgf8 gene regulatory network and the isthmic organizer: an evolutionary perspective
Open Access | Review | Published: 27 February 2025
Abstract
The midbrain-hindbrain boundary (MHB), also known as the isthmic organizer (IsO), plays a critical role in the developmental patterning of the posterior midbrain and anterior hindbrain. Understanding the wiring of this organizer’s deeply conserved gene regulatory network is of significant interest for both evolutionary and neurodevelopmental biology. Various secreted signalling molecules and transcription factors have been identified as being important components for the formation and function of the MHB. Among these, FGF8 is considered a primary mediator of IsO activity; it directs anterior-posterior patterning and promotes the specification and maintenance of the MHB. While the core gene regulatory network governing MHB development is well-characterized, the direct interactions between key regulatory genes and the cis-regulatory elements that control their spatiotemporal expression remain poorly understood. This review summarizes the current knowledge of the gene regulatory network underlying the formation of the vertebrate midbrain-hindbrain boundary. We focus in particular on Fgf8 and its regulatory landscape from an evolutionary perspective.
Keywords
Fgf8, midbrain-hindbrain boundary, gene regulatory networks, enhancer, regulatory landscape
Introduction
The vertebrate brain is a highly complex organ that develops from a simple sheet of epithelial cells known as the neural plate. During development, this initially simple structure becomes progressively more complex as the cells of the embryo divide, migrate, and acquire specific developmental identities. Neurulation transforms the neural plate into the neural tube through a multistep process that involves morphogenetic and inductive processes in which the brain is formed anteriorly and the spinal cord posteriorly. Subsequently, the brain undergoes division into three primary vesicles: the forebrain, midbrain, and hindbrain. Later in development the forebrain and hindbrain subdivide into secondary vesicles that eventually give rise to the adult brain structures.
The patterning underlying this spatial organization of the brain is directed by specialized signalling centres, so called organizers, which via morphogenetic signalling can pattern and induce cell fates in adjacent cells. The tripartite division of the brain can be traced back to ancestral chordates during evolution (Wada et al., 1998) and several of the secondary organizers coordinating vertebrate brain patterning have been proposed to have ancient deuterostome origins (Imai et al., 2009; Pani et al., 2012). Among them is the midbrain-hindbrain boundary (MHB), or isthmic organizer (IsO), which is essential for the developmental patterning of the posterior midbrain and anterior hindbrain (Wurst and Bally-Cuif, 2001). The deeply conserved MHB gene regulatory network (GRN) (Pani et al., 2012) makes its study interesting from an evolutionary perspective but it is also important to understand the underlying developmental mechanisms that can be disturbed in congenital disorders of the midbrain and hindbrain (Doherty et al., 2013; Gibbs et al., 2017).
Various secreted signalling molecules and transcription factors have been identified in the gene regulatory network that direct MHB specification and maintenance. Among them are Fibroblast growth factor 8 (Fgf8), members of the wnt-family of proteins (Wnt1, Wnt3, Wnt10b), as well as Paired homeobox 2/5/8 (Pax2/5/8), Engrailed 1/2 (En1/2) and Lim homeobox 1b (Lmx1b) transcription factors (Hidalgo-Sánchez et al., 2022; Wurst and Bally-Cuif, 2001). Several lines of evidence suggest that the main inductive molecule mediating IsO activity and directing anterior-posterior patterning in this region is FGF8. It is expressed in the MHB in all major vertebrate lineages (Christen and Slack, 1997; Crossley and Martin, 1995; Hidalgo-Sánchez et al., 1999a; Reifers et al., 1998). Furthermore, both loss-of-function experiments and ectopic expression experiments have demonstrated that it plays an essential role in the induction and maintenance of this region (Chi et al., 2003; Crossley et al., 1996; Irving and Mason, 2000; Jászai et al., 2003; Liu et al., 1999; Martinez et al., 1999; Meyers et al., 1998; Reifers et al., 1998; Sato and Joyner, 2009). Even though the core gene regulatory network directing MHB development is well characterized, and the essential role of Fgf8 in this system is well known, the direct interactions between key genes and their trans-acting regulatory factors are not well understood, nor is the evolution of the cis-regulatory elements that mediate their interactions. We will here review current knowledge on the MHB gene regulatory network with an emphasis on Fgf8 and its regulatory landscape from an evolutionary perspective.
Positioning, Activation and Maintenance of the Isthmic Organizer in vertebrates
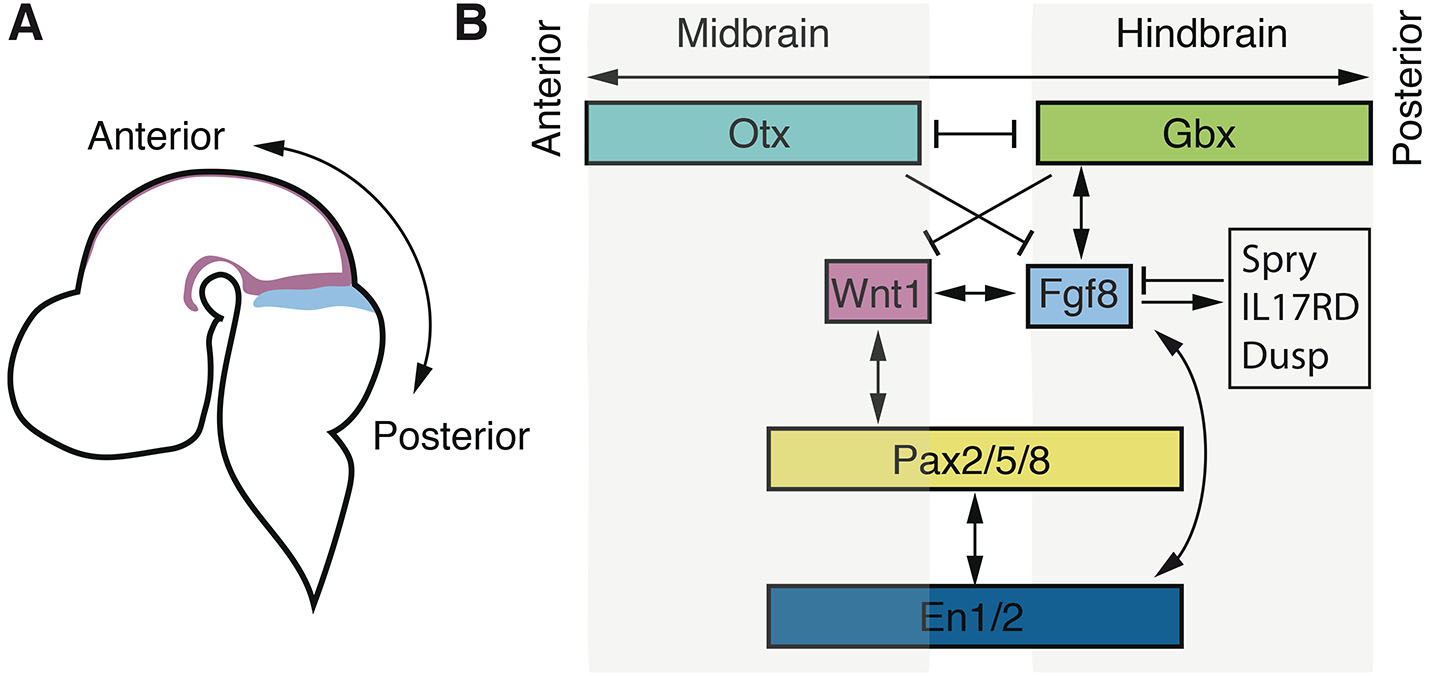
MHB formation involves multiple steps, including its positioning during gastrulation at the junction of Otx and Gbx expression, the establishment of the isthmus organizer activity through Fgf8 expression, and the maintenance of its regional identity and organizing activities (Gibbs et al., 2017; Wurst and Bally-Cuif, 2001). The development of the IsO is initiated within the neural plate as Otx and Gbx transcription factors are expressed. Otx2 expression is localised to the prospective forebrain and midbrain, and is required for specification of the most anterior parts of the brain including the secondary prosencephalon, diencephalon and the mesencephalon (Rhinn et al., 1998). Gbx2, on the other hand is expressed in the prospective hindbrain and is required for proper hindbrain development (Wassarman et al., 1997). The Gbx2 and Otx2 expression domains are slightly overlapping before a sharp boundary becomes evident; at the onset of Fgf8 MHB expression during early somitogenesis, these expression domains are adjacent but mutually exclusive (Fig. 1). The juxtaposition of Otx and Gbx gene expression is evolutionary conserved in vertebrates and is important for the positioning of the IsO in Xenopus (Glavic et al., 2002; Tour et al., 2002a; Tour et al., 2002b), zebrafish (Foucher et al., 2006; Mercier et al., 1995; Rhinn et al., 2003; Rhinn et al., 2009), chick (Garda et al., 2001; Katahira et al., 2000), and mouse (Broccoli et al., 1999; Li and Joyner, 2001; Martinez-Barbera et al., 2001; Millet et al., 1999). Other IsO-core genes also define the expression boundary of Otx and Gbx, suggesting a complex and dynamic process of MHB establishment (Martinez-Barbera et al., 2001; Rhinn et al., 2003). Still, genetic ablation of Gbx2 or Otx2 in mouse (Li and Joyner, 2001; Millet et al., 1999) and Gbx1/2 in zebrafish (Su et al., 2014) demonstrate that they are not required for the induction of Fgf8 expression. Thus, although not properly positioned, the MHB gene expression programme is initiated even in the absence of these transcription factors. Once the IsO is established and Fgf8 expression is induced, it requires the reciprocal repressive interaction of Otx2 and Gbx2 to be maintained (Li and Joyner, 2001; Rhinn et al., 2003; Su et al., 2014) (Fig. 1).
It has been proposed that several independent parallel signalling pathways are activated during the initial establishment of the MHB and that they involve Pax2, Wnt1, and Fgf8 respectively (Rhinn and Brand, 2001). However, bead implantation experiments and genetic ablation have shown that Fgf8 is the main inducer of posterior midbrain morphogenesis and anterior hindbrain patterning (Crossley et al., 1996; Meyers et al., 1998; Reifers et al., 1998; Shamim et al., 1999). Fgf8 ultimately induces tectum on its anterior side and cerebellum on its posterior side, while repressing the anterior-most hoxa2 expression in the anterior hindbrain (Irving and Mason, 2000; Sato and Joyner, 2009). The expression of Fgf8 is initiated at the border between the Otx and Gbx domains and localized to the anterior hindbrain region overlapping with the anterior most Gbx2 expression (Hidalgo-Sánchez et al., 1999b; Katahira et al., 2000). Despite the essential importance of Fgf8 expression in the IsO, no specific evolutionary conserved transcription factor has unambiguously been demonstrated to control induction of Fgf8 expression in the IsO directly.
In mice, genetic perturbation experiments suggested that the LIM homeobox transcription factor 1 beta (Lmx1b) and the paired-box family (Pax2/5/8) of transcription factors were important for initiation of Fgf8 expression (Guo et al., 2007; Ye et al., 2001). In mice, Lmx1b expression partially overlaps that of Fgf8 during initiation of the IsO. Analysis of Lmx1b-/- mice suggested the complete absence of Fgf8 expression in the MHB (Guo et al., 2007). However, recent data using radioactive in situ hybridisation demonstrate that Fgf8 expression is indeed induced in the MHB of these mice (Sherf et al., 2015). Also, double knockdown of Lmx1b.1 and Lmx1b.2 in zebrafish lead to progressive loss of Fgf8a only during the maintenance phase (O’Hara et al., 2005). Misexpression of Lmx1b in chick shows that Fgf8 is only induced in the adjacent surrounding cells (Matsunaga et al., 2002), further suggesting that any inductive effect of LMX1B on Fgf8 expression is only indirect.
Although Fgf8 MHB expression and associated anatomical structures are completely absent in Pax2 knockout mice in the C3H/He genetic background, MHB specification and development is normal in the C57Bl/6 background (Schwarz et al., 1997; Ye et al., 2001). Also, in zebrafish noi mutants that carry a functional deletion of Pax2a, expression of the core MHB genes is initiated and then progressively lost in the maintenance phase (Lun and Brand, 1998). Despite these discrepancies, Pax transcription factors could potentially be important inducers of Fgf8 gene expression since redundant function of other Pax transcription factor family members has been described (Schwarz et al., 1997) that in combination with genetic variation could buffer for induction of Fgf8 expression. Likewise, cooperativity and redundant function of distinct classes of trans-acting factors could potentially provide additional robustness during the induction of gene expression in the initiation phase. To further clarify this and identify direct regulators of Fgf8, genetic studies of compound mutants in combination with biochemical approaches are required.
Overall, previous studies have demonstrated an evolutionary conserved role in MHB development for several secreted molecules and transcription factors (Hidalgo-Sánchez et al., 2022; Wurst and Bally-Cuif, 2001). The induction of gene expression for these core factors is dependent on parallel signalling pathways (Canning et al., 2007; Lun and Brand, 1998) whereas the maintenance is characterized by extensive interdependent feedback loops (Dworkin and Jane, 2013; Gibbs et al., 2017) (Fig. 1). In most cases, perturbation of these core factors does not affect the initial establishment of the MHB but still lead to severe defects in the maintenance of the IsO and to later malformations in the midbrain-hindbrain region (McMahon et al., 1992; Lee et al., 1997; Adams et al., 2000; Lun et al., 1998; Hirata, 2001; Itoh et al., 2002; Buckles et al., 2004; Chung et al., 2006; Sherf et al., 2015). This observation suggests that distinct regulatory cues direct the initiation and maintenance of IsO gene expression and that its patterning activity relies on the interdependent expression of these core MHB factors, mediated via positive and negative regulatory feedback loops (Hidalgo-Sánchez et al., 2022; Wurst and Bally-Cuif, 2001) (Fig. 1). Thus, genetic studies in various vertebrate model organisms have led to a model of MHB development that includes positioning, initiation, and maintenance, and that involves a highly conserved core gene regulatory network but for which the initial inductive molecular cascade and direct regulatory events remain unknown.
Evolution of the midbrain-hindbrain boundary
During early development, the vertebrate brain is divided into three regions that give rise to the forebrain, the midbrain and the hindbrain. This tripartite regional organisation of the brain is thought to have ancient origin, predating the evolution of the chordate lineage (Lowe et al., 2003; Wada et al., 1998) and has been suggested to have been present in the last common urbilaterian ancestor (Hirth et al., 2003; Urbach, 2007). Although there is a partial overlap between genes demarcating the tripartite subdivision of the brain, and those in the vertebrate MHB gene regulatory network, the origin and evolution of the MHB as an organizer is less well defined (Holland, 2015).
In Drosophila melanogaster, Otx and Gbx orthologues are expressed in a juxtaposed pattern similar to that of vertebrates. The boundary between the anteriorly expressed Otd (Otx) and the more posteriorly expressed Unpg (Gbx) aligns with the deutocerebral–tritocerebral boundary (DTB), which has been hypothesized to correspond to the vertebrate MHB (Bridi et al., 2020; Hirth et al., 2003; Urbach, 2007). Similar to vertebrates, the D. melanogaster orthologues of Wnt1 and En are expressed in the vicinity of the Otd-Unpg interface. Initial reports describing the expression of Fgf8-orthologues did not support a role in Drosophila boundary formation (Hirth et al., 2003; Urbach, 2007), but more recent data show that the Fgf8-orthologues Ths and Pyr are expressed in the DTB. Genetic experiments indicate that downregulation of these genes or the FGF8-receptor Htl leads to altered expression of En and Unpg (Bridi et al., 2020). Although this suggests that FGF8 has an ancient role associated with boundary formation in the tripartite brain, it does not appear to have the organizing activity intrinsic to the vertebrate IsO (Bridi et al., 2020).
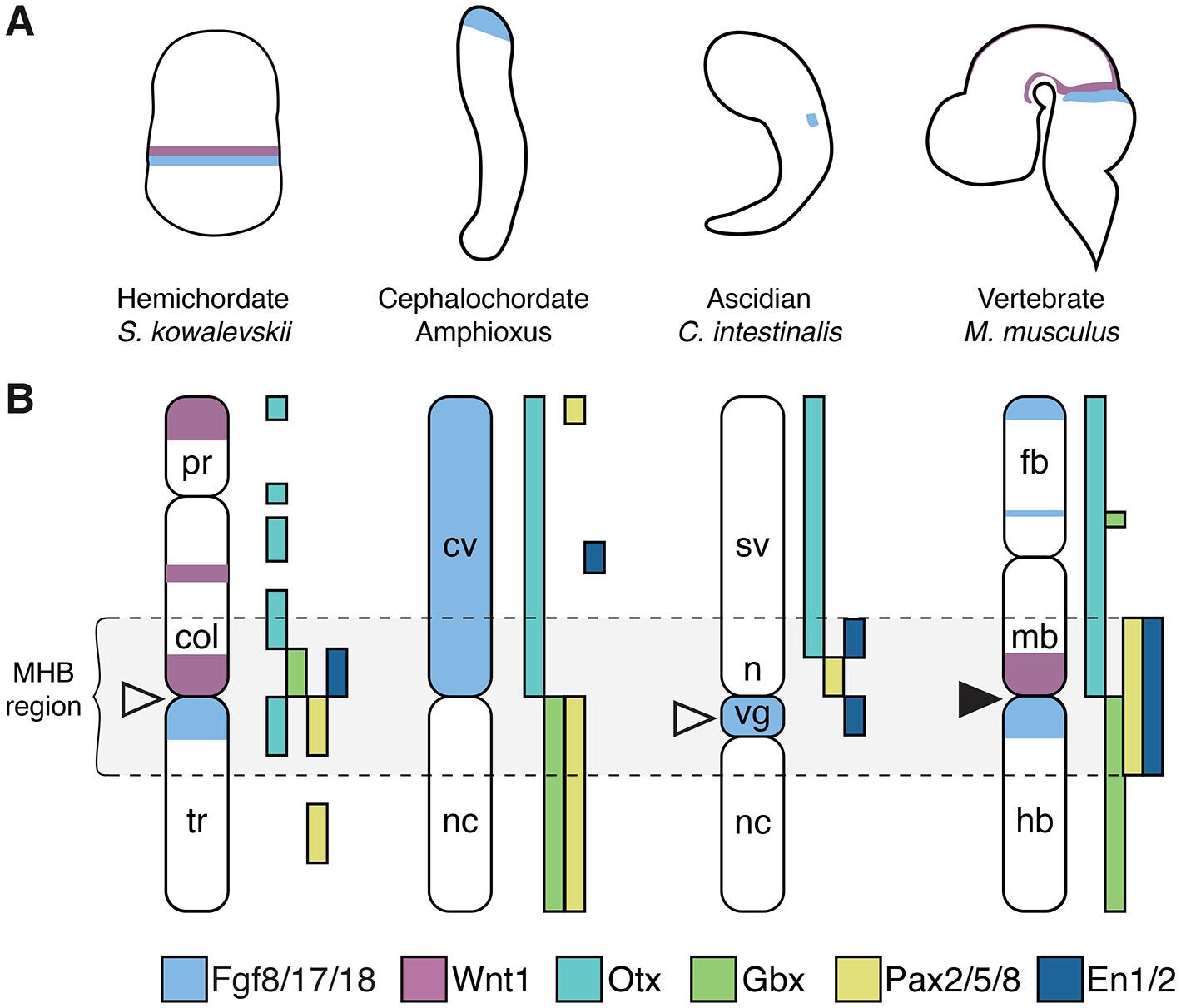
In the hemichordate S. kowalevskii, a gene expression programme reminiscent of that in the vertebrate MHB is present in the developing ectoderm (Lowe et al., 2003; Pani et al., 2012). Importantly, the juxtaposed expression pattern of homologues to Wnt1 and the key MHB inducer Fgf8 (Fgf8/17/18) is localized to adjacent domains at the collar–trunk coelom boundary, respectively (Fig. 2). Similar juxtaposition is also evident in the case of the transcription factors Otx and Gbx, while Pax2/5/8 and En are also expressed in the region (Fig. 2). However, Otx-Gbx expression is reversed as compared to Fgf8-Wnt1 expression in S. kowalevskii: Otx expression overlap with that of Fgf8 while Gbx expression overlap with that of Wnt1. Also distinct to vertebrates, the Pax2/5/8 and En expression domains do not overlap. Still, based on these data, and those from experiments in which FGF and WNT signalling was perturbed, it has been suggested that the gene regulatory network directing the MHB secondary organizer has ancient deuterostome origin pre-dating vertebrate evolution (Pani et al., 2012).
Fig. 2. Chordate expression of midbrain-hindbrain boundary (MHB) genes.
(A) The anatomical context shown in (B). (B) A summary of the gene expression patterns of Fgf8, Wnt1, Otx, Gbx, Pax and En homologues in the ectoderm of S. kowalevskii, amphioxus, and C. intestinalis, and in the brain of M. musculus. col: collar, cv: cerebral vesicle, fb: forebrain, hb: hindbrain, mb: midbrain, n: neck, nc: nerve cord, pr: proboscis, sv: sensory vesicle, tr: trunk, vg, visceral ganglion. White arrowheads indicate the presumptive IsO in S. kowalevskii and C. intestinalis; the black arrowhead indicates the vertebrate IsO. The diagrams are not drawn to scale.
In the cephalochordate amphioxus, Otx and Gbx genes are expressed in juxtaposition at the border between the cerebral vesicle and the hindbrain in a region corresponding to the MHB (Castro et al., 2006). However, other genes involved in vertebrate IsO specification display distinct expression patterns: Fgf8/17/18 is expressed in the entire cerebral vesicle and Wnt1 is not expressed in the region (Bertrand et al., 2011; Holland, 2013), while Pax2/5/8 is expressed throughout the hindbrain (Holland, 2013; Kozmik et al., 1999) (Fig. 2). This suggests that amphioxus have some of the genetic machinery required for specification of the IsO but that the key molecules mediating organizing activity in the vertebrate IsO are lacking and their cross-regulatory interactions are not present (Holland, 2015).
Data from the urochordate C. intestinalis show that expression of the Fgf8/17/18 orthologue is localized to the brain larval visceral ganglion at the tailbud stage, juxtaposed to Pax2/5/8 expression and (as it is in the vertebrate IsO) co-expressed with En (Imai et al., 2002; Imai et al., 2009) (Fig. 2). Knockdown of Fgf8/17/18 leads to loss of Pax2/5/8 expression and expansion of Otx and FoxB, which are both expressed in the anterior central nervous system domain, adjacent to Pax2/5/8 (Imai et al., 2009). This change resembles the transformation of the anterior hindbrain into an expanded midbrain that occurs in vertebrates in Fgf8 loss of function mutants (Chi et al., 2003; Reifers et al., 1998). Thus, although it has been suggested that this Fgf8/17/18 expression could also correspond to the more posterior expression in rhombomere 4 in zebrafish (Cañestro et al., 2005), it appears that Fgf8/17/18 in C. intestinalis mediates organizing activity that is important for midbrain-hindbrain regionalisation, which in turn suggests that it corresponds at least to a partial IsO homologous to that in the vertebrate MHB (Imai et al., 2009).
Still, in vertebrates, the positioning and maintenance of the Fgf8 expression in the IsO depends on the combined cross-regulatory interactions of Wnt1, Otx, Gbx, Pax2/5/8, En1/2, among others. While En expression overlap with Fgf8/17/18 in C. intestinalis, the expression domain of Pax2/5/8 is located more anteriorly, and Wnt1 and Gbx are not present in the C. intestinalis genome (Dehal et al., 2002; Hino et al., 2002; Wada et al., 2003) (Fig. 2). Furthermore, in contrast to vertebrates but similar to the regulation of the Drosophila Fgf8 homologues Ths and Pyr (Stathopoulos et al., 2004), the expression of C. intestinalis Fgf8/17/18 is restricted by the transcriptional repressor Snail (Imai et al., 2009). These studies indicate that while some of the genetic components of the IsO existed before chordate evolution, the complete cross-regulatory interactions that induce and maintain the vertebrate IsO likely evolved after the divergence of the cephalochordates and olfactores lineages. While the GRN that patterns the MHB appears to have an ancient origin, it is not yet fully established to what extent the neural circuits arising from this region in invertebrates perform functions comparable to those in vertebrates. Still, in D. melanogaster, the brain region derived from the DTB integrates sensorimotor information and mediates balance and motor coordination. This may imply that the vertebrate MHB originated from an ancestral region already characterized by the presence of neural circuits related to balance and vestibular function (Bridi et al., 2020). Additionally, co-expression of En and tyrosine hydroxylase in the S. kowalevskii collar-trunk coelom boundary indicates that these cells may be homologous to vertebrate midbrain dopaminergic neurons.
In none of the species that display IsO-resembling Fgf8 gene expression, have the upstream transcription factors and the regulatory events that lead to its initiation been reported, and it remains unclear how the regulation of Fgf8 evolved. Nevertheless, despite variations in the genetic networks specifying the MHB, the findings from these evolutionarily distant species suggest that Fgf8 had an ancestral role in boundary formation that later evolved to include the organizing activity that is crucial for the vertebrate IsO.
The FGF protein family and signalling
The Fibroblast Growth Factor (FGF) protein family is a group of structurally related growth factors that are crucial in numerous biological processes. During embryonic development they are important for cell proliferation, differentiation, migration, and survival in multiple organs, making them essential for tissue patterning and morphogenesis. In the MHB region, in addition to induction and maintenance of the MHB GRN, FGF-signalling have multiple functions, including cell survival (Basson et al., 2008), maintenance of symmetrical proliferative divisions in the midbrain ventricular zone (Lahti et al., 2011), and axonal guidance (Irving et al., 2002). Based on phylogenetic analyses in humans and mice, 22 FGF members are divided into seven subfamilies — Fgf1, Fgf4, Fgf7, Fgf8, Fgf9, Fgf11, and Fgf15/19 (Ornitz and Itoh, 2022). These subfamilies are further classified into three functional groups: canonical, hormone-like, and intracellular FGFs. Canonical FGFs, which include the FGF8 as well as the FGF1, FGF4, FGF7, and FGF9 subfamilies, are primarily secreted ligands that bind tightly to heparan sulphate proteoglycans, which regulate their interactions with specific FGF receptors (Mohammadi et al., 2005; Yayon et al., 1991).
In zebrafish, the genome contains 27 Fgf genes, with all seven subfamilies represented and two paralogues for some genes (including Fgf8) due to the genome duplication that occurred after the teleost split (Itoh and Konishi, 2007). The expansion of FGF genes likely occurred in two major phases, first after the separation of the protostome and deuterostome lineages and then a second expansion in early vertebrate evolution (Itoh and Ornitz, 2011). Reflecting this time frame, the C. intestinalis, amphioxus and S. kowalevskii genomes harbour 6, 8 and 6 Fgf genes, respectively (Bertrand et al., 2011; Oulion et al., 2012; Satou et al., 2002), while D. melanogaster and the nematode C. elegans have 3 FGF genes. Members of the FGF8 sub-family are present in all of these species and have been described in various arthropods (Oulion et al., 2012).
FGF-signalling is mediated by four distinct high-affinity receptor tyrosine kinases in vertebrates, originating from one single gene in primitive chordates. Although FGFR1, FGFR2, and FGFR3 are all expressed in the embryonic midbrain in mice, only FGFR1 expression overlaps with that of Fgf8 at the IsO (Blak et al., 2005), and mice lacking FGFR1 expression manifest a more severe MHB phenotype than mice lacking FGFR2 or FGFR3 (Blak et al., 2007; Trokovic et al., 2003). Still, FGFR2 and FGFR3 are also thought to be important transducers of the IsO signal, and act redundantly in conjunction with FGFR1 to regulate the production of neuronal cells such as dopaminergic neurons in the ventral midbrain, as well as promoting survival of dorsal neuroectoderm (Saarimäki-Vire et al., 2007). In concordance with this idea, redundant function of FGFR1 and FGFR2 has also been shown in the zebrafish (Leerberg et al., 2019). The activation of canonical FGFs triggers four major signalling pathways: RAS-MAPK, PI3-AKT, PLCγ/protein kinase C, and STAT pathways (Ornitz and Itoh, 2015), and there is evidence suggesting FGF also signals via nuclear FGFR localization (Förthmann et al., 2015).
FGF-signalling in the MHB region induces various genes that modulate the pathway, many of which that are members of a Fgf8 syn-expression group that is well conserved in vertebrates (Eblaghie et al., 2003; Fürthauer et al., 2002; Haines et al., 2006; Hirate and Okamoto, 2006; Li et al., 2007; Lin et al., 2002; Lin et al., 2005; Minowada et al., 1999; Tsang et al., 2002; Tsang et al., 2004). Positive regulators include the Canopy FGF signalling regulator 1 (Cnpy1) and the transmembrane protein FLRT3, which enhances FGF signalling by promoting FGFR maturation in the endoplasmic reticulum (Hirate and Okamoto, 2006), and by direct interaction with the FGFR1 receptor (Böttcher et al., 2004; Haines et al., 2006), respectively. Negative regulators include members of the Sprouty (SPRY) (Yu et al., 2011) and the Dual Specificity Phosphatase (DUSP) (Li et al., 2007) family of proteins, as well as the transmembrane protein Interleukin 17 receptor D (IL17RD) (Lin et al., 2005). SPRY1 and SPRY2 inhibits the RAS-MAPK pathway and regulates PI3K-AKT signalling (Ornitz and Itoh, 2015) while DUSP6 attenuates FGF signalling by dephosphorylation of MAPK (Camps et al., 1998). L17RD blocks the nuclear translocation of activated MAPK and may also inhibit FGF signalling directly interacting with FGFR1 (Fürthauer et al., 2002; Torii et al., 2004; Tsang et al., 2002).
The final transcriptional output of FGF-signalling is mediated by nuclear effectors including the ETV4 and ETV5 transcription factors of the ETS transcription factor family, which regulate the expression of target genes such as Dusp6 (Ekerot et al., 2008; Znosko et al., 2010). Activation of both positive and negative regulators of FGF signalling is not yet fully understood but may be critical for regulating the timing of active FGF signalling, fine tuning expression patterns, and shaping the FGF signalling gradient in developing tissues, including the MHB.
The role of FGF8 in the IsO
FGF8 is a canonical FGF, and the founding member of a subfamily consisting of FGF8, FGF17, and FGF18 in mice and humans. The zebrafish has 6 members of this family, as the genome contain two paralogous genes for FGF8 (Fgf8a and Fgf8b) and FGF18 (Fgf18a and Fgf18b), one gene encoding FGF17, and the additional Fgf24 gene that has been lost in the tetrapod lineage. The Fgf8 gene subfamily members have highly dynamic patterns of gene expression during vertebrate development but several of their expression domains are overlapping, including in the MHB (Maruoka et al., 1998; Ohuchi et al., 2000). Still, several lines of evidence suggest that Fgf8 is the key inducer of IsO activity.
Initial experiments in chick demonstrated that insertion of FGF8-soaked beads in the midbrain exerts similar polarising activity as the IsO, and induces ectopic expression of the MHB markers En2, Pax2, and Wnt1 in the midbrain (Crossley et al., 1996; Irving and Mason, 2000; Martinez et al., 1999). In mice, Fgf8 gene expression in the MHB begins at 4-5 somites, i. e. at an earlier stage than that of Fgf17 and Fgf18, and it is required for the latter´s expression in the region (Chi et al., 2003; Liu et al., 2003). In zebrafish, Fgf8a is expressed in the anterior hindbrain at the late gastrula stage, and this expression gradually splits into three anterior rhombomeres, r1, r2, and r4 at early somitogenesis (Reifers et al., 1998). Similar to the situation in mice, zebrafish Fgf8a is the most strongly expressed paralogue in the early MHB, while Fgf8b, Fgf18b and Fgf24 expression begins slightly later (Jovelin et al., 2010). In mice, removal of Fgf8 gene expression in the IsO leads to the downregulation of genes in the MHB gene regulatory network, with subsequent progressive aplasia of the posterior midbrain and anterior hindbrain structures (Chi et al., 2003; Meyers et al., 1998), while Fgf17 and Fgf18 mutants show mild and no phenotypes, respectively (Liu et al., 2002; Xu et al., 2000). Although the severe phenotype of Fgf8 MHB knockout mice is caused by increased cell death, moderately reduced levels of FGF-signalling result in changes in cell fate specification (Basson et al., 2008), and zebrafish Fgf8a mutants lack cerebellar tissue not because of cell death but transformation of the cerebellar region into midbrain cells (Jászai et al., 2003; Picker et al., 1999; Reifers et al., 1998; Tallafuß and Bally-Cuif, 2003). Thus, Fgf8 not only affects cell survival but also functions to specify regional identity. The differences in apoptosis between species may be because of species-specific redundancy of other survival factors, e.g., zebrafish WNTs (Buckles et al., 2004). In addition to anteroposterior patterning, Fgf8-mediated signalling has been suggested to contribute to the dorsoventral patterning of the MHB in zebrafish and medaka (Carl and Wittbrodt, 1999; Fürthauer et al., 1997), and in mice dorsal MHB structures are more sensitive to reduction of Fgf8 expression (Chi et al., 2003; Meyers et al., 1998). Thus, these studies have demonstrated the importance of FGF8 for IsO activity and for the polarized gene expression patterns that underlie regionalisation and cell fate choices in the MHB.
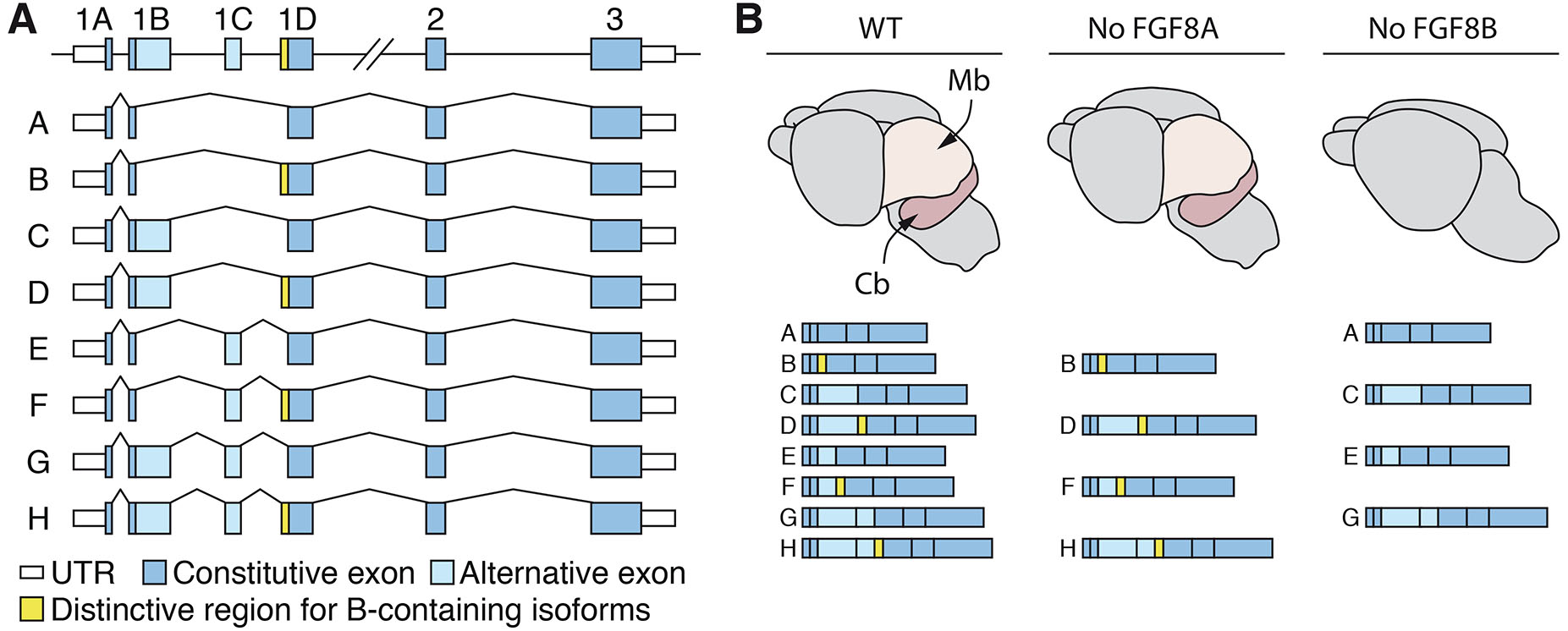
Adding to the complexity of FGF8-signalling in the MHB region, multiple isoforms have been identified. In chicken, Xenopus and zebrafish, two Fgf8 isoforms have been described, while eight have been reported in mice, and four in humans (Sunmonu et al., 2011) (Fig. 3A). The two most conserved isoforms, Fgf8A and Fgf8B, which are present from fish to mammals, exert differential activity in the MHB. While FGF8A regulates midbrain formation (Lee et al., 1997; Liu et al., 1999; Sato et al., 2001), FGF8B also promotes development of the cerebellum (Liu et al., 1999; Sato et al., 2001). A potential explanation for this discrepancy is the higher receptor binding affinity of FGF8B isoform due to the presence of phenylalanine 32 in the N-terminal region (Olsen et al., 2006). Indeed, in both chicken and zebrafish, FGF8A and FGF8B activate FGF signalling with different intensity, and reduced concentrations of Fgf8b can mimic the effect of Fgf8a (Inoue et al., 2006; Sato et al., 2001). However, even at very high concentrations, electroporation of Fgf8a cannot mimic FGF8B activity and induce cerebellar development (Fletcher et al., 2006; Sunmonu et al., 2011), and in both mouse and chicken only FGF8B can ectopically induce hindbrain genes such as Gbx2 in the midbrain region (Liu et al., 1999; Sato et al., 2001). In addition, it was shown that only FGF8B misexpression activates the Ras–Erk pathway in the midbrain of the chick (Sato and Nakamura, 2004). These results suggest that differences in signalling properties are both quantitative and qualitative, and this interpretation is further supported by data from mouse genetic experiments. Removal of all Fgf8B-containing isoforms in mice phenocopies the complete deletion of Fgf8, leading to downregulation of key IsO regulatory genes, and subsequent loss of the MHB-derived structures (Guo et al., 2010) (Fig. 3B). In contrast, even though removal of Fgf8A isoforms leads to growth retardation and perinatal lethality, MHB patterning and growth are not affected, including when overall FGF-signalling is reduced by the additional removal of Fgf17 expression (Guo et al., 2010) (Fig. 3B). Thus, these data demonstrated that diverse Fgf8 isoforms exert distinct signalling strength, but also highlighted likely differences in the downstream signal transduction cascade and established that Fgf8b is the main isoform mediating IsO activity in vivo. Despite them having different signalling properties and apparently distinct roles in specific tissues, there is little, if any, differential regulation of Fgf8 isoforms during development either temporal or spatially: multiple isoforms are expressed simultaneously in different tissues (Blunt et al., 1997). Still, the spatiotemporal regulation of Fgf8 gene expression is tightly controlled in the vertebrate embryo, including in the MHB.
Fig. 3. Differential requirement for Fgf8 isoforms in the midbrain-hindbrain boundary (MHB).
(A) Representation of all Fgf8 isoforms reported in the mouse. Isoforms C, D, G, and H are not present in humans due to a premature stop codon in exon 1B. Exon 1C is only present in placental mammals, while Fgf8A and Fgf8B are evolutionarily conserved across vertebrates, from fish to mammals. (B) Schematic illustration of e18.5 brains from wildtype controls, mutant mice with a splice-acceptor mutation that removes all Fgf8A-containing isoforms (middle), or with a splice-acceptor mutation that remove all Fgf8B-containing isoforms (right). Only the removal of Fgf8B-containing isoforms results in the loss of posterior midbrain structures and cerebellar aplasia, mimicking the phenotype observed in MHB Fgf8 KO mice. Cb: cerebellum; Mb: midbrain.
The regulatory landscape of Fgf8
In vertebrates, the precise expression of many developmental genes, including Fgf8, is regulated by multiple cis-regulatory elements. Beyond the promoter regions near the target genes, distant transcriptional enhancers play a crucial role in activating the specific temporal and spatial expression patterns of these genes (Long et al., 2016). Although these enhancers can be located within or beyond neighbouring genes and influence gene expression across large genomic distances (Lettice et al., 2003), physical proximity between enhancers and their target promoters is crucial for gene activation (Chen et al., 2024; Zuin et al., 2022). In most animals, the genome is partitioned into so-called topologically associated domains (TADs) (Acemel and Lupiáñez, 2023), which consist of segmental chromosomal regions that are primarily self-interacting (Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012). TADs largely overlap with gene regulatory domains (Symmons et al., 2014) and facilitate enhancer-promoter interactions by increasing the probability of physical contact.
Concurrently, TADs limit the genomic range in which enhancers can act and thus reduce the probability of ectopic activation of non-target genes. This restriction imposes evolutionary constraints, as disrupting this organization can result in gene misexpression and cause severe developmental malformations or disease (Flavahan et al., 2016; Franke et al., 2016; Lupiáñez et al., 2015; Rajderkar et al., 2023; Symmons et al., 2016). Consequently, TADs are more often reorganized as intact modules during genome evolution (Vietri Rudan et al., 2015; Farré et al., 2015; Li et al., 2022).
Similarly, since regulatory sequences can be located in neighbouring genes or even further away, cis-regulatory constraints contribute to shaping conservation of syntenic regions (Irimia et al., 2012; Kikuta et al., 2007). Ancient conservation of microsynteny has been used to identify genomic regulatory blocks (GRBs) that are regions of the genome containing developmental genes that are physically linked to nearby bystander genes because they are part of their gene regulatory landscape (Irimia et al., 2012; Kikuta et al., 2007). The boundaries of GRBs are predictive for TAD boundaries and GRBs tend to overlap with TADs (Harmston et al., 2017).
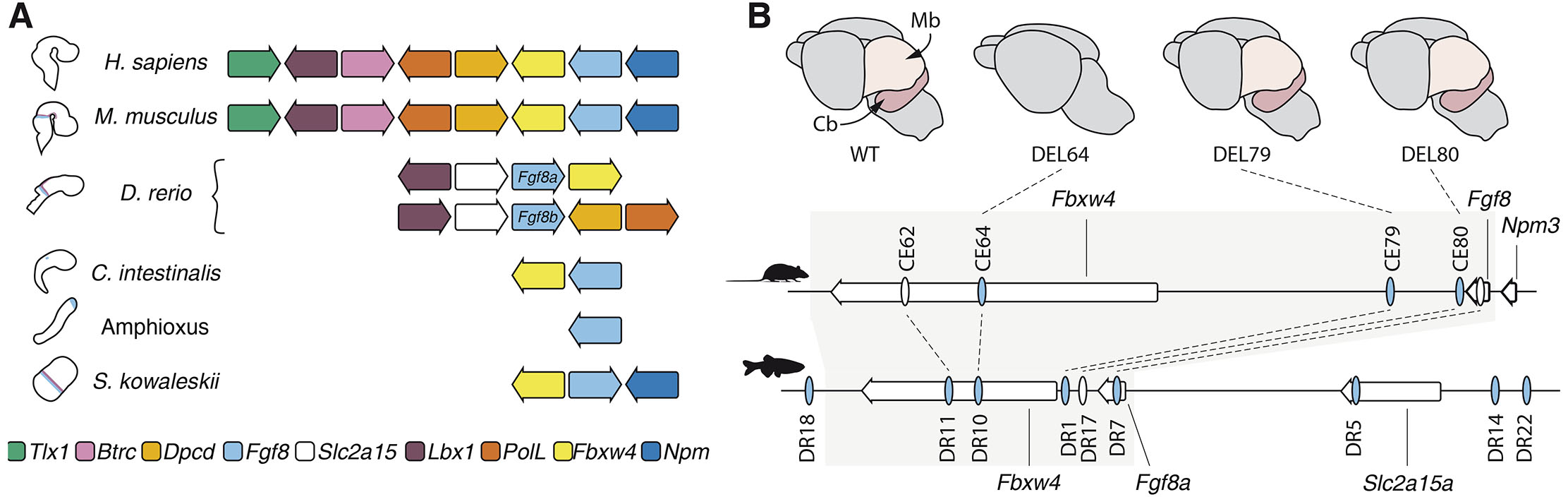
In mice and humans, Fgf8 is located in a syntenic region that also contains Tlx1, Lbx1, Btrc, Poll, Dpcd, Fbxw4, and Npm3 (Fig. 4A). In zebrafish Btrc, Poll, Dpcd and Npm3 genes have been lost from the locus, while the Fgf8a/Fbxw4 gene pair is inverted and the Slc2a15a gene intercalates between Lbx1 and Fbxw4 (Fig. 4A). The microsynteny of the Fgf8/Fbxw4 gene pair is ancient and conserved in both the hemichordate S. kowalevskii (Simakov et al., 2015) and the urochordate C. intestinalis (Jovelin et al., 2010) suggesting that they form an ancient GRB (Cañestro et al., 2007) (Fig. 4A). Corroborating this regulatory linkage, in mice Fbxw4 and Fgf8 are the only genes that are located within the same TAD, and this region also largely overlaps the Fgf8 regulatory domain (Cova et al., 2023; Marinić et al., 2013). Given the fundamental importance of Fgf8 in chordate development, one could speculate that distant regulatory elements directing Fgf8 gene expression were present within the Fbxw4 gene already in very early chordates, and that loss of the Fgf8/Fbxw4 synteny in amphioxus is related to the complete lack of an IsO in this species (Fig. 4A).
Fig. 4. The regulatory landscape of Fgf8 is embedded within an ancient gene regulatory block.
(A) Representation of the synteny of the Fgf8-containing genomic region in deuterostome species. The Fbxw4-Fgf8 gene pair is retained in all species with an IsO-like expression of Fgf8. (B) Putative midbrain-hindbrain boundary (MHB) enhancers (ovals) in the Fgf8 genomic region in mouse and zebrafish. Blue indicates reported MHB expression in transgenic assays. Grey box depicts the Fbxw4-Fgf8 syntenic region that corresponds to the mouse topologically associated domains (TAD) and that is conserved in the deuterostome lineage. The upper row illustrates e18.5 mouse brains: wildtype (left), CE64 enhancer deletion mutants (middle left), CE79 mutants (middle right), and CE80 mutants (right). The removal of CE64 results in complete absence of Fgf8 expression in the MHB, and subsequent loss of the posterior midbrain and cerebellum. Cb: cerebellum; Mb: midbrain.
The enhancer landscape of the Fgf8 locus has been extensively studied in both mice and zebrafish, and several putative enhancers with tissue-specific regulatory activity have been identified (Beermann et al., 2006; Hörnblad et al., 2021; Hu et al., 2004; Inoue et al., 2006; Komisarczuk et al., 2009; Marinić et al., 2013; Sasaki et al., 2008). Most of these putative enhancers are located downstream of the Fgf8 gene, in the intergenic region between Fbxw4 and Fgf8, or embedded within the Fbxw4 gene. These enhancers frequently have overlapping tissue-specific regulatory activity so that multiple elements can drive expression in similar spatio-temporal domains (Marinić et al., 2013). Such redundancy confers robustness to gene expression and allows for evolution of regulatory novelty (Frankel et al., 2010; Hong et al., 2008; Hörnblad et al., 2021; Osterwalder et al., 2018; Perry et al., 2010)
A large reporter screen of human conserved elements identified three putative MHB enhancers (CE80, CE79, and CE64) that could drive reporter expression in the MHB region of mice at e10.5 (Marinić et al., 2013). Two of these elements, CE80 and CE79, are located in a region proximal to the 3’ end of the Fgf8 gene and had been identified previously in a similar screen (Beermann et al., 2006) (Fig. 4B). The distal CE64 is located 120kb downstream of Fgf8 in the 4th intron of Fbxw4 (Fig. 4B). All of these putative enhancers are highly conserved from fish to humans, and were also identified as putative enhancers in the zebrafish (Inoue et al., 2006; Komisarczuk et al., 2009). However, in zebrafish, only CE79 and CE64 showed regulatory activity in the MHB in reporter assays, while CE80 was active in other tissues (Fig. 4B). In addition, five other putative MHB enhancers were also reported in zebrafish (Komisarczuk et al., 2009) (Fig. 4B). Importantly, in vivo enhancer deletions demonstrated that in the mouse only the most distal CE64 MHB enhancer is essential for Fgf8 expression; its removal results in complete failure to induce Fgf8 expression in the MHB, with subsequent cerebellar aplasia and loss of posterior midbrain structures (Hörnblad et al., 2021) (Fig. 4B). In contrast, single deletions of the more proximal CE80 and CE79, or deletion of both together caused only a very small reduction in Fgf8 expression when CE80 was absent, and animals lacking C79 or CE80 (or both) developed normally (Hörnblad et al., 2021) (Fig. 4B). Although these experiments do not exclude the possibility that the CE80 and CE79 enhancers may function redundantly with CE64 in the MHB maintenance phase, they established CE64 as an essential main enhancer required for both initiation and maintenance of Fgf8 expression in the IsO. In zebrafish, functional data on the in vivo importance of putative enhancers are incomplete, but detailed transgenic reporter experiments have shown that the zebrafish equivalent to CE79 cannot recapitulate the very early expression of Fgf8a in the MHB (Inoue et al., 2008). This observation suggests that, similar to the mouse, CE79 has a less important role for initiation of Fgf8a expression. Interestingly, the synteny of Fgf8/Fbxw4 has been lost in the Fgf8b locus, and this paralogue only becomes expressed after Fgf8a in the MHB. This may indicate that enhancers within the Fbxw4 gene are important for initiation of Fgf8a expression, also in zebrafish. Taken together, these data highlight the need for functional in vivo characterization of putative regulatory elements in several species to fully understand the complexity of the Fgf8 regulatory logic in the MHB and how it has evolved.
Many conserved genetic interactions in the core MHB gene regulatory network have been described, mainly in the positioning and maintenance of the IsO. Less is known about the induction of the MHB, and the direct regulatory interactions between transcription factors and regulatory elements at specific loci that initiate and maintain the MHB gene expression programme. Although it was reported that PAX2 can bind to CE79 in zebrafish (Inoue et al., 2008), and phylogenetic footprinting together with in vivo functional dissection of the mouse CE64 enhancer could identify essential regulatory motifs (Hörnblad et al., 2021), no additional data have demonstrated the direct interaction between trans-acting transcription factors and their cognate Fgf8 enhancers in the MHB.
Discussion
Fgf8 signalling emanating from the IsO coordinates the patterning, differentiation, and growth of the vertebrate MHB region. This makes the IsO a prime model to explore how localised gene expression drives large-scale developmental processes, thereby ensuring stereotypic patterning and morphogenesis during embryonic development. The ancient origin of the gene regulatory networks that direct the formation and function of the MHB-region also makes it a valuable model for studying evolutionary conservation and plasticity in gene regulation.
A wealth of studies has identified a network of genes involved in regulating IsO formation, maintenance and function, and the morphogenetic processes that accompany these gene expression programmes have also been described. Less is known about the cell-type specific, genome-wide, and direct regulatory interactions, which occur in the context of the topological organisation of the genome. Thus, the direct transcriptional regulators of Fgf8 in the MHB are not known, and a full understanding of the cis-regulatory sequences through which they act is still missing. A critical step will therefore be to identify these transcription factors and to understand their specific roles in initiating and maintaining Fgf8 MHB gene expression. Identification of the transcription factors will help in the decoding of the complex regulatory logic of Fgf8 and reveal how the GRN have been evolutionarily conserved and adapted across vertebrates.
While key enhancers have been functionally validated in mice, more comprehensive in vivo functional characterization across diverse species is required to unravel the evolutionary dynamics of Fgf8 regulation. Despite the sequence conservation of putative cis-regulatory elements from zebrafish to human, the temporal activity of these enhancers and their interactions with trans-acting factors may differ across species. Furthermore, it remains to be understood whether the deep conservation of these putative enhancers reflects a rigid ‘enhanceosome’-like regulatory architecture or whether their organisation is more flexible and allows for reshuffling of regulatory motifs according to the ´billboard’ model (Kulkarni and Arnosti, 2003; Thanos and Maniatis, 1995). In mice, the presence of one essential MHB enhancer containing both redundant and non-redundant regulatory features suggests a mixture of the two (Hörnblad et al., 2021). Similar cis-regulatory architecture could potentially hold true for other species such as zebrafish, but the high number of putative enhancers driving reporter expression in the MHB in this species may also indicate a distinct, and more distributed regulatory architecture. Still, it is clear that the regulatory potential of a developmental enhancer, as demonstrated by transgenic reporter assays, does not necessarily reflect its relative importance in the tissue in vivo.
The implementation of CRISPR-based enhancer deletions and the generation of highly precise transgenic reporter lines will help in the exploration of the spatiotemporal control of key MHB genes, including Fgf8. Integrating these genetic tools with innovative techniques, such as chromatin conformation capture and single-cell sequencing technology, will help to unravel the wiring of the MHB gene regulatory network, and in particular the interactions between trans-acting factors and their target cis-regulatory elements. By exploring the genome-wide regulatory landscapes and gene expression profiles of distinct cell types at the MHB, as well as identifying the transcription factor-enhancer interactions that direct midbrain and hindbrain development, we can gain significant insights into the molecular orchestration of vertebrate brain patterning, development and evolution.
References
Acemel R. D., Lupiáñez D. G. (2023). Evolution of 3D chromatin organization at different scales. Current Opinion in Genetics & Development 78: 102019.
Adams K. A., Maida J. M., Golden J. A., Riddle R. D. (2000). The transcription factor Lmx1b maintains Wnt1 expression within the isthmic organizer. Development 127: 1857-1867.
Basson M. A., Echevarria D., Petersen Ahn C., Sudarov A., Joyner A. L., Mason I. J., Martinez S., Martin G. R. (2008). Specific regions within the embryonic midbrain and cerebellum require different levels of FGF signaling during development. Development 135: 889-898.
Beermann F., Kaloulis K., Hofmann D., Murisier F., Bucher P., Trumpp A. (2006). Identification of evolutionarily conserved regulatory elements in the mouse Fgf8 locus. genesis 44: 1-6.
Bertrand S., Camasses A., Somorjai I., Belgacem M. R., Chabrol O., Escande M.L., Pontarotti P., Escriva H. (2011). Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proceedings of the National Academy of Sciences 108: 9160-9165.
Blak A. A., Naserke T., Saarimäki-Vire J., Peltopuro P., Giraldo-Velasquez M., Vogt Weisenhorn D. M., Prakash N., Sendtner M., Partanen J., Wurst W. (2007). Fgfr2 and Fgfr3 are not required for patterning and maintenance of the midbrain and anterior hindbrain. Developmental Biology 303: 231-243.
Blak A. A., Naserke T., Weisenhorn D. M. V., Prakash N., Partanen J., Wurst W. (2005). Expression of Fgf receptors 1, 2, and 3 in the developing mid‐ and hindbrain of the mouse. Developmental Dynamics 233: 1023-1030.
Blunt A. G., Lawshé A., Cunningham M. L., Seto M. L., Ornitz D. M., MacArthur C. A. (1997). Overlapping Expression and Redundant Activation of Mesenchymal Fibroblast Growth Factor (FGF) Receptors by Alternatively Spliced FGF-8 Ligands. Journal of Biological Chemistry 272: 3733-3738.
Böttcher R. T., Pollet N., Delius H., Niehrs C. (2004). The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nature Cell Biology 6: 38-44.
Bridi J. C., Ludlow Z. N., Kottler B., Hartmann B., Vanden Broeck L., Dearlove J., Göker M., Strausfeld N. J., Callaerts P., Hirth F. (2020). Ancestral regulatory mechanisms specify conserved midbrain circuitry in arthropods and vertebrates. Proceedings of the National Academy of Sciences 117: 19544-19555.
Broccoli V., Boncinelli E., Wurst W. (1999). The caudal limit of Otx2 expression positions the isthmic organizer. Nature 401: 164-168.
Buckles G. R., Thorpe C. J., Ramel M.C., Lekven A. C. (2004). Combinatorial Wnt control of zebrafish midbrain–hindbrain boundary formation. Mechanisms of Development 121: 437-447.
Camps M., Nichols A., Gillieron C., Antonsson B., Muda M., Chabert C., Boschert U., Arkinstall S. (1998). Catalytic Activation of the Phosphatase MKP-3 by ERK2 Mitogen-Activated Protein Kinase. Science 280: 1262-1265.
Cañestro C., Bassham S., Postlethwait J. (2005). Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain. Developmental Biology 285: 298-315.
Cañestro C., Yokoi H., Postlethwait J. H. (2007). Evolutionary developmental biology and genomics. Nature Reviews Genetics 8: 932-942.
Canning C. A., Lee L., Irving C., Mason I., Jones C. M. (2007). Sustained interactive Wnt and FGF signaling is required to maintain isthmic identity. Developmental Biology 305: 276-286.
Carl M., Wittbrodt J. (1999). Graded interference with FGF signalling reveals its dorsoventral asymmetry at the mid-hindbrain boundary. Development 126: 5659-5667.
Castro L. F. C., Rasmussen S. L.K., Holland P. W.H., Holland N. D., Holland L. Z. (2006). A Gbx homeobox gene in amphioxus: Insights into ancestry of the ANTP class and evolution of the midbrain/hindbrain boundary. Developmental Biology 295: 40-51.
Chen Z., Snetkova V., Bower G., Jacinto S., Clock B., Dizehchi A., Barozzi I., Mannion B. J., Alcaina-Caro A., Lopez-Rios J., Dickel D. E., Visel A., Pennacchio L. A., Kvon E. Z. (2024). Increased enhancer–promoter interactions during developmental enhancer activation in mammals. Nature Genetics 56: 675-685.
Chi C. L., Martinez S., Wurst W., Martin G. R. (2003). The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development 130: 2633-2644.
Chung A. C., Xu X., Niederreither K. A., Cooney A. J., (2006). Loss of orphan nuclear receptor GCNF function disrupts forebrain development and the establishment of the isthmic organizer. Developmental Biology 293: 13-24.
Christen B., Slack J. M.W. (1997). FGF-8Is Associated with Anteroposterior Patterning and Limb Regeneration inXenopus. Developmental Biology 192: 455-466.
Cova G., Glaser J., Schöpflin R., Prada-Medina C. A., Ali S., Franke M., Falcone R., Federer M., Ponzi E., Ficarella R., Novara F., Wittler L., Timmermann B., Gentile M., Zuffardi O., Spielmann M., Mundlos S. (2023). Combinatorial effects on gene expression at the Lbx1/Fgf8 locus resolve split-hand/foot malformation type 3. Nature Communications 14: 1475.
Crossley P. H., Martin G. R. (1995). The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121: 439-451.
Crossley P. H., Martinez S., Martin G. R. (1996). Midbrain development induced by FGF8 in the chick embryo. Nature 380: 66-68.
Dehal P., Satou Y., Campbell R. K., Chapman J., Degnan B., Tomaso A.D., Davidson B., De Gregorio M., Gelpke M., Goodstein D. M., Harafuji N., Hastings K. E. M., (2002). The Draft Genome of Ciona intestinalis: Insights into Chordate and Vertebrate Origins. Science 298: 2157-2167.
Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376-380.
Doherty D., Millen K. J., Barkovich A. J. (2013). Midbrain and hindbrain malformations: advances in clinical diagnosis, imaging, and genetics. The Lancet Neurology 12: 381-393.
Dworkin S., Jane S. M. (2013). Novel mechanisms that pattern and shape the midbrain-hindbrain boundary. Cellular and Molecular Life Sciences 70: 3365-3374.
Eblaghie M. C., Lunn J.S., Dickinson R. J., Münsterberg A. E., Sanz-Ezquerro J.J., Farrell E. R., Mathers J., Keyse S. M., Storey K., Tickle C. (2003). Negative Feedback Regulation of FGF Signaling Levels by Pyst1/MKP3 in Chick Embryos. Current Biology 13: 1009-1018.
Ekerot M., Stavridis M. P., Delavaine L., Mitchell M. P., Staples C., Owens D. M., Keenan I. D., Dickinson R. J., Storey K. G., Keyse S. M. (2008). Negative-feedback regulation of FGF signalling by DUSP6/MKP-3 is driven by ERK1/2 and mediated by Ets factor binding to a conserved site within the DUSP6 / MKP - 3 gene promoter. Biochemical Journal 412: 287-298.
Farré M., Robinson T. J., Ruiz‐Herrera A. (2015). An Integrative Breakage Model of genome architecture, reshuffling and evolution. BioEssays 37: 479-488.
Flavahan W. A., Drier Y., Liau B. B., Gillespie S. M., Venteicher A. S., Stemmer-Rachamimov A. O., Suvà M. L., Bernstein B. E. (2016). Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110-114.
Fletcher R. B., Baker J. C., Harland R. M. (2006). FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 133: 1703-1714.
Förthmann B., Aletta J. M., Lee Y.W., Terranova C., Birkaya B., Stachowiak E. K., Stachowiak M. K., Claus P. (2015). Coalition of Nuclear Receptors in the Nervous System. Journal of Cellular Physiology 230: 2875-2880.
Foucher I., Mione M., Simeone A., Acampora D., Bally-Cuif L., Houart C. (2006). Differentiation of cerebellar cell identities in absence of Fgf signalling in zebrafish Otx morphants. Development 133: 1891-1900.
Franke M., Ibrahim D. M., Andrey G., Schwarzer W., Heinrich V., Schöpflin R., Kraft K., Kempfer R., Jerković I., Chan W.L., Spielmann M., Timmermann B., Wittler L., Kurth I., Cambiaso P., Zuffardi O., Houge G., Lambie L., Brancati F., Pombo A., Vingron M., Spitz F., Mundlos S. (2016). Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538: 265-269.
Frankel N., Davis G. K., Vargas D., Wang S., Payre F., Stern D. L. (2010). Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490-493.
Fürthauer M., Lin W., Ang S.L., Thisse B., Thisse C. (2002). Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nature Cell Biology 4: 170-174.
Fürthauer M., Thisse C., Thisse B. (1997). A role for FGF-8 in the dorsoventral patterning of the zebrafish gastrula. Development 124: 4253-4264.
Garda A.L., Echevarrı́a D., Martı́nez S. (2001). Neuroepithelial co-expression of Gbx2 and Otx2 precedes Fgf8 expression in the isthmic organizer. Mechanisms of Development 101: 111-118.
Gibbs H. C., Chang-Gonzalez A., Hwang W., Yeh A. T., Lekven A. C. (2017). Midbrain-Hindbrain Boundary Morphogenesis: At the Intersection of Wnt and Fgf Signaling. Frontiers in Neuroanatomy 11: 64.
Glavic A., Gómez-Skarmeta J. L., Mayor R. (2002). The homeoprotein Xiro1 is required for midbrain-hindbrain boundary formation. Development 129: 1609-1621.
Guo C., Qiu H.Y., Huang Y., Chen H., Yang R.Q., Chen S.D., Johnson R. L., Chen Z.F., Ding Y.Q. (2007). Lmx1b is essential for Fgf8 and Wnt1 expression in the isthmic organizer during tectum and cerebellum development in mice. Development 134: 317-325.
Guo Q., Li K., Sunmonu N. A., Li J. Y.H. (2010). Fgf8b-containing spliceforms, but not Fgf8a, are essential for Fgf8 function during development of the midbrain and cerebellum. Developmental Biology 338: 183-192.
Haines B. P., Wheldon L. M., Summerbell D., Heath J. K., Rigby P. W.J. (2006). Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Developmental Biology 297: 14-25.
Harmston N., Ing-Simmons E., Tan G., Perry M., Merkenschlager M., Lenhard B. (2017). Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nature Communications 8: 441.
Hidalgo-Sánchez M., Andreu-Cervera A., Villa-Carballar S., Echevarria D. (2022). An Update on the Molecular Mechanism of the Vertebrate Isthmic Organizer Development in the Context of the Neuromeric Model. Frontiers in Neuroanatomy 16: 826976.
Hidalgo-Sánchez M., Millet S., Simeone A., Alvarado-Mallart R.-M. (1999a). Comparative analysis of Otx2, Gbx2, Pax2, Fgf8 and Wnt1 gene expressions during the formation of the chick midbrain/hindbrain domain. Mechanisms of Development 81: 175-178.
Hidalgo-Sánchez M., Simeone A., Alvarado-Mallart R. M. (1999b). Fgf8 and Gbx2 induction concomitant with Otx2 repression is correlated with midbrain-hindbrain fate of caudal prosencephalon. Development 126: 3191-3203.
Hino K., Satou Y., Yagi K., Satoh N. (2002). A genomewide survey of developmentally relevant genes in Ciona intestinalis. Development Genes and Evolution 213: 264-272.
Hirata H. (2001). Hes1 and Hes3 regulate maintenance of the isthmic organizer and development of the mid/hindbrain. The EMBO Journal 20: 4454-4466.
Hirate Y., Okamoto H. (2006). Canopy1, a Novel Regulator of FGF Signaling around the Midbrain-Hindbrain Boundary in Zebrafish. Current Biology 16: 421-427.
Hirth F., Kammermeier L., Frei E., Walldorf U., Noll M., Reichert H. (2003). An urbilaterian origin of the tripartite brain: developmental genetic insights from Drosophila. Development 130: 2365-2373.
Holland L. Z. (2013). Evolution of new characters after whole genome duplications: Insights from amphioxus. Seminars in Cell & Developmental Biology 24: 101-109.
Holland L. Z. (2015). The origin and evolution of chordate nervous systems. Philosophical Transactions of the Royal Society B: Biological Sciences 370: 20150048.
Hong J.W., Hendrix D. A., Levine M. S. (2008). Shadow Enhancers as a Source of Evolutionary Novelty. Science 321: 1314-1314.
Hörnblad A., Bastide S., Langenfeld K., Langa F., Spitz F. (2021). Dissection of the Fgf8 regulatory landscape by in vivo CRISPR-editing reveals extensive intra- and inter-enhancer redundancy. Nature Communications 12: 439.
Hu T., Yamagishi H., Maeda J., McAnally J., Yamagishi C., Srivastava D. (2004). Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development 131: 5491-5502.
Imai K. S., Satoh N., Satou Y. (2002). Region specific gene expressions in the central nervous system of the ascidian embryo. Mechanisms of Development 119: S275-S277.
Imai K. S., Stolfi A., Levine M., Satou Y. (2009). Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136: 285-293.
Inoue F., Nagayoshi S., Ota S., Islam M. E., Tonou‐Fujimori N., Odaira Y., Kawakami K., Yamasu K. (2006). Genomic organization, alternative splicing, and multiple regulatory regions of the zebrafish fgf8 gene. Development, Growth & Differentiation 48: 447-462.
Inoue F., Parvin M. S., Yamasu K. (2008). Transcription of fgf8 is regulated by activating and repressive cis-elements at the midbrain–hindbrain boundary in zebrafish embryos. Developmental Biology 316: 471-486.
Irimia M., Tena J. J., Alexis M. S., Fernandez-Miñan A., Maeso I., Bogdanović O., de la Calle-Mustienes E., Roy S. W., Gómez-Skarmeta J. L., Fraser H. B. (2012). Extensive conservation of ancient microsynteny across metazoans due to cis -regulatory constraints. Genome Research 22: 2356-2367.
Irving C., Malhas A., Guthrie S., Mason I. (2002). Establishing the trochlear motor axon trajectory: role of the isthmic organiser and Fgf8. Development 129: 5389-5398.
Irving C., Mason I. (2000). Signalling by FGF8 from the isthmus patterns anterior hindbrain and establishes the anterior limit of Hox gene expression. Development 127: 177-186.
Itoh M., Kudoh T., Dedekian M., Kim C.H., Chitnis A. B. (2002). A role for iro1 and iro7 in the establishment of an anteroposterior compartment of the ectoderm adjacent to the midbrain-hindbrain boundary. Development 129: 2317-2327.
Itoh N., Konishi M. (2007). The Zebrafish fgf Family. Zebrafish 4: 179-186.
Itoh N., Ornitz D. M. (2011). Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. Journal of Biochemistry 149: 121-130.
Jászai J., Reifers F., Picker A., Langenberg T., Brand M. (2003). Isthmus-to-midbrain transformation in the absence of midbrain-hindbrain organizer activity. Development 130: 6611-6623.
Jovelin R., Yan Y.L., He X., Catchen J., Amores A., Canestro C., Yokoi H., Postlethwait J. H. (2010). Evolution of developmental regulation in the vertebrate FgfD subfamily. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 314B: 33-56.
Katahira T., Sato T., Sugiyama S., Okafuji T., Araki I., Funahashi J., Nakamura H. (2000). Interaction between Otx2 and Gbx2 defines the organizing center for the optic tectum. Mechanisms of Development 91: 43-52.
Kikuta H., Laplante M., Navratilova P., Komisarczuk A. Z., Engström P. G., Fredman D., Akalin A., Caccamo M., Sealy I., Howe K., Ghislain J., Pezeron G., Mourrain P., Ellingsen S., Oates A. C., Thisse C., Thisse B., Foucher I., Adolf B., Geling A., Lenhard B., Becker T. S. (2007). Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Research 17: 545-555.
Komisarczuk A. Z., Kawakami K., Becker T. S. (2009). Cis-regulation and chromosomal rearrangement of the fgf8 locus after the teleost/tetrapod split. Developmental Biology 336: 301-312.
Kozmik Z., Holland N. D., Kalousova A., Paces J., Schubert M., Holland L. Z. (1999). Characterization of an amphioxus paired box gene, AmphiPax2/5/8 : developmental expression patterns in optic support cells, nephridium, thyroid-like structures and pharyngeal gill slits, but not in the midbrain-hindbrain boundary region. Development 126: 1295-1304.
Kulkarni M. M., Arnosti D. N. (2003). Information display by transcriptional enhancers. Development 130: 6569-6575.
Lahti L., Saarimäki-Vire J., Rita H., Partanen J. (2011). FGF signaling gradient maintains symmetrical proliferative divisions of midbrain neuronal progenitors. Developmental Biology 349: 270-282.
Lee S. M. K., Danielian P. S., Fritzsch B., McMahon A. P. (1997). Evidence that FGF8 signalling from the midbrain-hindbrain junction regulates growth and polarity in the developing midbrain. Development 124: 959-969.
Leerberg D. M., Hopton R. E., Draper B. W. (2019). Fibroblast Growth Factor Receptors Function Redundantly During Zebrafish Embryonic Development. Genetics 212: 1301-1319.
Lettice L. A. (2003). A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Human Molecular Genetics 12: 1725-1735.
Li C., Scott D. A., Hatch E., Tian X., Mansour S. L. (2007). Dusp6 ( Mkp3 ) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development 134: 167-176.
Li D., He M., Tang Q., Tian S., Zhang J., Li Y., Wang D., Jin L., Ning C., Zhu W., Hu S., Long K., Ma J., Liu J., Zhang Z., Li M. (2022). Comparative 3D genome architecture in vertebrates. BMC Biology 20: 99.
Li J. Y. H., Joyner A. L. (2001). Otx2 and Gbx2 are required for refinement and not induction of mid-hindbrain gene expression. Development 128: 4979-4991.
Lin W., Fürthauer M., Thisse B., Thisse C., Jing N., Ang S.L. (2002). Cloning of the mouse Sef gene and comparative analysis of its expression with Fgf8 and Spry2 during embryogenesis. Mechanisms of Development 113: 163-168.
Lin W., Jing N., Basson M. A., Dierich A., Licht J., Ang S.L. (2005). Synergistic activity of Sef and Sprouty proteins in regulating the expression ofGbx2 in the mid-hindbrain region. genesis 41: 110-115.
Liu A., Li J. Y. H., Bromleigh C., Lao Z., Niswander L. A., Joyner A. L. (2003). FGF17b and FGF18 have different midbrain regulatory properties from FGF8b or activated FGF receptors. Development 130: 6175-6185.
Liu A., Losos K., Joyner A. L. (1999). FGF8 can activate Gbx2 and transform regions of the rostral mouse brain into a hindbrain fate. Development 126: 4827-4838.
Liu Z., Xu J., Colvin J. S., Ornitz D. M. (2002). Coordination of chondrogenesis and osteogenesis by fibroblast growth factor 18. Genes & Development 16: 859-869.
Long H. K., Prescott S. L., Wysocka J. (2016). Ever-Changing Landscapes: Transcriptional Enhancers in Development and Evolution. Cell 167: 1170-1187.
Lowe C. J., Wu M., Salic A., Evans L., Lander E., Stange-Thomann N., Gruber C. E., Gerhart J., Kirschner M. (2003). Anteroposterior Patterning in Hemichordates and the Origins of the Chordate Nervous System. Cell 113: 853-865.
Lun K., Brand M. (1998). A series of no isthmus ( noi ) alleles of the zebrafish pax2 . 1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development 125: 3049-3062.
Lupiáñez D. G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J. M., Laxova R., Santos-Simarro F., Gilbert-Dussardier B., Wittler L., Borschiwer M., Haas S. A., Osterwalder M., Franke M., Timmermann B., Hecht J., Spielmann M., Visel A., Mundlos S. (2015). Disruptions of Topological Chromatin Domains Cause Pathogenic Rewiring of Gene-Enhancer Interactions. Cell 161: 1012-1025.
Marinić M., Aktas T., Ruf S., Spitz F. (2013). An Integrated Holo-Enhancer Unit Defines Tissue and Gene Specificity of the Fgf8 Regulatory Landscape. Developmental Cell 24: 530-542.
Martinez S., Crossley P. H., Cobos I., Rubenstein J. L. R., Martin G. R. (1999). FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development 126: 1189-1200.
Martinez-Barbera J. P., Signore M., Boyl P. P., Puelles E., Acampora D., Gogoi R., Schubert F., Lumsden A., Simeone A. (2001). Regionalisation of anterior neuroectoderm and its competence in responding to forebrain and midbrain inducing activities depend on mutual antagonism between OTX2 and GBX2. Development 128: 4789-4800.
Maruoka Y., Ohbayashi N., Hoshikawa M., Itoh N., Hogan B. L.M., Furuta Y. (1998). Comparison of the expression of three highly related genes, Fgf8, Fgf17 and Fgf18, in the mouse embryo. Mechanisms of Development 74: 175-177.
Matsunaga E., Katahira T., Nakamura H. (2002). Role of Lmx1b and Wnt1 in mesencephalon and metencephalon development. Development 129: 5269-5277.
McMahon A. P., Joyner A. L., Bradley A., McMahon J. A. (1992). The midbrain-hindbrain phenotype of Wnt-1−Wnt-1− mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69: 581-595.
Mercier P., Simeone A., Cotelli F., Boncinelli E., (1995). Expression pattern of two otx genes suggests a role in specifying anterior body structures in zebrafish. The International journal of developmental biology 39: 559-573.
Meyers E. N., Lewandoski M., Martin G. R. (1998). An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nature Genetics 18: 136-141.
Millet S., Campbell K., Epstein D. J., Losos K., Harris E., Joyner A. L. (1999). A role for Gbx2 in repression of Otx2 and positioning the mid/hindbrain organizer. Nature 401: 161-164.
Minowada G., Jarvis L. A., Chi C. L., Neubüser A., Sun X., Hacohen N., Krasnow M. A., Martin G. R. (1999). Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 126: 4465-4475.
Mohammadi M., Olsen S. K., Goetz R. (2005). A protein canyon in the FGF–FGF receptor dimer selects from an à la carte menu of heparan sulfate motifs. Current Opinion in Structural Biology 15: 506-516.
Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N. L., Meisig J., Sedat J., Gribnau J., Barillot E., Blüthgen N., Dekker J., Heard E. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381-385.
O'Hara F. P., Beck E., Barr L. K., Wong L. L., Kessler D. S., Riddle R. D. (2005). Zebrafish Lmx1b.1 and Lmx1b.2 are required for maintenance of the isthmic organizer. Development 132: 3163-3173.
Ohuchi H., Kimura S., Watamoto M., Itoh N. (2000). Involvement of fibroblast growth factor (FGF)18-FGF8 signaling in specification of left-right asymmetry and brain and limb development of the chick embryo. Mechanisms of Development 95: 55-66.
Olsen S. K., Li J. Y.H., Bromleigh C., Eliseenkova A. V., Ibrahimi O. A., Lao Z., Zhang F., Linhardt R. J., Joyner A. L., Mohammadi M. (2006). Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes & Development 20: 185-198.
Ornitz D. M., Itoh N. (2022). New developments in the biology of fibroblast growth factors. WIREs Mechanisms of Disease 14: e1549.
Ornitz D. M., Itoh N. (2015). The Fibroblast Growth Factor signaling pathway. WIREs Developmental Biology 4: 215-266.
Osterwalder M., Barozzi I., Tissières V., Fukuda-Yuzawa Y., Mannion B. J., Afzal S. Y., Lee E. A., Zhu Y., Plajzer-Frick I., Pickle C. S., Kato M., Garvin T. H., Pham Q. T., Harrington A. N., Akiyama J. A., Afzal V., Lopez-Rios J., Dickel D. E., Visel A., Pennacchio L. A. (2018). Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554: 239-243.
Oulion S., Bertrand S., Escriva H. (2012). Evolution of the FGF Gene Family. International Journal of Evolutionary Biology 2012: 1-12.
Pani A. M., Mullarkey E. E., Aronowicz J., Assimacopoulos S., Grove E. A., Lowe C. J. (2012). Ancient deuterostome origins of vertebrate brain signalling centres. Nature 483: 289-294.
Perry M. W., Boettiger A. N., Bothma J. P., Levine M. (2010). Shadow Enhancers Foster Robustness of Drosophila Gastrulation. Current Biology 20: 1562-1567.
Picker A., Brennan C., Reifers F., Clarke J. D. W., Holder N., Brand M. (1999). Requirement for the zebrafish mid-hindbrain boundary in midbrain polarisation, mapping and confinement of the retinotectal projection*. Development 126: 2967-2978.
Rajderkar S., Barozzi I., Zhu Y., Hu R., Zhang Y., Li B., Alcaina Caro A., Fukuda-Yuzawa Y., Kelman G., Akeza A., Blow M. J., Pham Q., Harrington A. N., Godoy J., Meky E. M., von Maydell K., Hunter R. D., Akiyama J. A., Novak C. S., Plajzer-Frick I., Afzal V., Tran S., Lopez-Rios J., Talkowski M. E., Lloyd K. C. K., Ren B., Dickel D. E., Visel A., Pennacchio L. A. (2023). Topologically associating domain boundaries are required for normal genome function. Communications Biology 6: 435.
Reifers F., Böhli H., Walsh E. C., Crossley P. H., Stainier D. Y. R., Brand M. (1998). Fgf8 is mutated in zebrafish acerebellar ( ace ) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesisy. Development 125: 2381-2395.
Rhinn M., Brand M. (2001). The midbrain–hindbrain boundary organizer. Current Opinion in Neurobiology 11: 34-42.
Rhinn M., Dierich A., Shawlot W., Behringer R. R., Meur M. L., Ang S.L. (1998). Sequential roles for Otx2 in visceral endoderm and neuroectoderm for forebrain and midbrain induction and specification. Development 125: 845-856.
Rhinn M., Lun K., Ahrendt R., Geffarth M., Brand M. (2009). Zebrafish gbx1 refines the Midbrain-Hindbrain Boundary border and mediates the Wnt8 posteriorization signal. Neural Development 4: 12.
Rhinn M., Lun K., Amores A., Yan Y.L., Postlethwait J. H., Brand M. (2003). Cloning, expression and relationship of zebrafish gbx1 and gbx2 genes to Fgf signaling. Mechanisms of Development 120: 919-936.
Saarimäki-Vire J., Peltopuro P., Lahti L., Naserke T., Blak A. A., Vogt Weisenhorn D. M., Yu K., Ornitz D. M., Wurst W., Partanen J. (2007). Fibroblast Growth Factor Receptors Cooperate to Regulate Neural Progenitor Properties in the Developing Midbrain and Hindbrain. The Journal of Neuroscience 27: 8581-8592.
Sagai T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T. (2005). Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132: 797-803.
Sasaki T., Nishihara H., Hirakawa M., Fujimura K., Tanaka M., Kokubo N., Kimura-Yoshida C., Matsuo I., Sumiyama K., Saitou N., Shimogori T., Okada N. (2008). Possible involvement of SINEs in mammalian-specific brain formation. Proceedings of the National Academy of Sciences 105: 4220-4225.
Sato T., Araki I., Nakamura H. (2001). Inductive signal and tissue responsiveness defining the tectum and the cerebellum. Development 128: 2461-2469.
Sato T., Joyner A. L. (2009). The duration of Fgf8 isthmic organizer expression is key to patterning different tectal-isthmo-cerebellum structures. Development 136: 3617-3626.
Sato T., Nakamura H. (2004). The Fgf8 signal causes cerebellar differentiation by activating the Ras-ERK signaling pathway. Development 131: 4275-4285.
Satou Y., Imai K. S., Satoh N. (2002). Fgf genes in the basal chordate Ciona intestinalis. Development Genes and Evolution 212: 432-438.
Schwarz M., Alvarez-Bolado G., Urbánek P., Busslinger M., Gruss P. (1997). Conserved biological function between Pax- 2 and Pax- 5 in midbrain and cerebellum development: Evidence from targeted mutations. Proceedings of the National Academy of Sciences 94: 14518-14523.
Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. (2012). Three-Dimensional Folding and Functional Organization Principles of the Drosophila Genome. Cell 148: 458-472.
Shamim H., Mahmood R., Logan C., Doherty P., Lumsden A., Mason I. (1999). Sequential roles for Fgf4, En1 and Fgf8 in specification and regionalisation of the midbrain. Development 126: 945-959.
Sherf O., Nashelsky Zolotov L., Liser K., Tilleman H., Jovanovic V. M., Zega K., Jukic M. M., Brodski C. (2015). Otx2 Requires Lmx1b to Control the Development of Mesodiencephalic Dopaminergic Neurons. PLOS ONE 10: e0139697.
Simakov O., Kawashima T., Marlétaz F., Jenkins J., Koyanagi R., Mitros T., Hisata K., Bredeson J., Shoguchi E., Gyoja F., Yue J.X., Chen Y.C., Freeman R. M., Sasaki A., Hikosaka-Katayama T., Sato A., Fujie M., Baughman K. W., Levine J., Gonzalez P., Cameron C., Fritzenwanker J. H., Pani A. M., Goto H., Kanda M., Arakaki N., Yamasaki S., Qu J., Cree A., Ding Y., Dinh H. H., Dugan S., Holder M., Jhangiani S. N., Kovar C. L., Lee S. L., Lewis L. R., Morton D., Nazareth L. V., Okwuonu G., Santibanez J., Chen R., Richards S., Muzny D. M., Gillis A., Peshkin L., Wu M., Humphreys T., Su Y.H., Putnam N. H., Schmutz J., Fujiyama A., Yu J.K., Tagawa K., Worley K. C., Gibbs R. A., Kirschner M. W., Lowe C. J., Satoh N., Rokhsar D. S., Gerhart J. (2015). Hemichordate genomes and deuterostome origins. Nature 527: 459-465.
Stathopoulos A., Tam B., Ronshaugen M., Frasch M., Levine M. (2004). pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes & Development 18: 687-699.
Su C.Y., Kemp H. A., Moens C. B. (2014). Cerebellar development in the absence of Gbx function in zebrafish. Developmental Biology 386: 181-190.
Sunmonu N. A., Li K., Li J. Y.H. (2011). Numerous isoforms of Fgf8 reflect its multiple roles in the developing brain. Journal of Cellular Physiology 226: 1722-1726.
Symmons O., Pan L., Remeseiro S., Aktas T., Klein F., Huber W., Spitz F. (2016). The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Developmental Cell 39: 529-543.
Symmons O., Uslu V. V., Tsujimura T., Ruf S., Nassari S., Schwarzer W., Ettwiller L., Spitz F. (2014). Functional and topological characteristics of mammalian regulatory domains. Genome Research 24: 390-400.
Tallafuß A., Bally-Cuif L. (2003). Tracing of her5 progeny in zebrafish transgenics reveals the dynamics of midbrain-hindbrain neurogenesis and maintenance. Development 130: 4307-4323.
Thanos D., Maniatis T. (1995). Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell 83: 1091-1100.
Torii S., Kusakabe M., Yamamoto T., Maekawa M., Nishida E. (2004). Sef Is a Spatial Regulator for Ras/MAP Kinase Signaling. Developmental Cell 7: 33-44.
Tour E., Pillemer G., Gruenbaum Y., Fainsod A. (2002a). Gbx2 interacts with Otx2 and patterns the anterior–posterior axis during gastrulation in Xenopus. Mechanisms of Development 112: 141-151.
Tour E., Pillemer G., Gruenbaum Y., Fainsod A. (2002b). Otx2 can activate the isthmic organizer genetic network in the Xenopus embryo. Mechanisms of Development 110: 3-13.
Trokovic R. (2003). FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. The EMBO Journal 22: 1811-1823.
Tsang M., Friesel R., Kudoh T., Dawid I. B. (2002). Identification of Sef, a novel modulator of FGF signalling. Nature Cell Biology 4: 165-169.
Tsang M., Maegawa S., Kiang A., Habas R., Weinberg E., Dawid I. B. (2004). A role for MKP3 in axial patterning of the zebrafish embryo. Development 131: 2769-2779.
Urbach R. (2007). A procephalic territory in Drosophila exhibiting similarities and dissimilarities compared to the vertebrate midbrain/hindbrain boundary region. Neural Development 2: 23.
Urbánek P., Fetka I., Meisler M. H., Busslinger M. (1997). Cooperation of Pax2 and Pax5 in midbrain and cerebellum development. Proceedings of the National Academy of Sciences 94: 5703-5708.
Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D. T., Tanay A., Hadjur S., (2015). Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Reports 10: 1297-1309.
Wada H., Saiga H., Satoh N., Holland P. W. H. (1998). Tripartite organization of the ancestral chordate brain and the antiquity of placodes: insights from ascidian Pax-2/5/8, Hox and Otx genes. Development 125: 1113-1122.
Wada S., Tokuoka M., Shoguchi E., Kobayashi K., Di Gregorio A., Spagnuolo A., Branno M., Kohara Y., Rokhsar D., Levine M., Saiga H., Satoh N., Satou Y. (2003). A genomewide survey of developmentally relevant genes in Ciona intestinalis. Development Genes and Evolution 213: 222-234.
Wassarman K. M., Lewandoski M., Campbell K., Joyner A. L., Rubenstein J. L. R., Martinez S., Martin G. R. (1997). Specification of the anterior hindbrain and establishment of a normal mid/hindbrain organizer is dependent on Gbx2 gene function. Development 124: 2923-2934.
Wurst W., Bally-Cuif L. (2001). Neural plate patterning: Upstream and downstream of the isthmic organizer. Nature Reviews Neuroscience 2: 99-108.
Xu J., Liu Z., Ornitz D. M. (2000). Temporal and spatial gradients of Fgf8 and Fgf17 regulate proliferation and differentiation of midline cerebellar structures. Development 127: 1833-1843.
Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991). Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64: 841-848.
Ye W., Bouchard M., Stone D., Liu X., Vella F., Lee J., Nakamura H., Ang S.L., Busslinger M., Rosenthal A. (2001). Distinct regulators control the expression of the mid-hindbrain organizer signal FGF8. Nature Neuroscience 4: 1175-1181.
Yu T., Yaguchi Y., Echevarria D., Martinez S., Basson M. A. (2011). Sprouty genes prevent excessive FGF signalling in multiple cell types throughout development of the cerebellum. Development 138: 2957-2968.
Znosko W. A., Yu S., Thomas K., Molina G. A., Li C., Tsang W., Dawid I. B., Moon A. M., Tsang M. (2010). Overlapping functions of Pea3 ETS transcription factors in FGF signaling during zebrafish development. Developmental Biology 342: 11-25.
Zuin J., Roth G., Zhan Y., Cramard J., Redolfi J., Piskadlo E., Mach P., Kryzhanovska M., Tihanyi G., Kohler H., Eder M., Leemans C., van Steensel B., Meister P., Smallwood S., Giorgetti L. (2022). Nonlinear control of transcription through enhancer–promoter interactions. Nature 604: 571-577.