Int. J. Dev. Biol. 68: 117 - 126 (2024)
Histological characterisation of the horn bud region in 58 day old bovine fetuses
Open Access | Original Article | Published: 14 August 2024
Abstract
The presence of horns in domestic ruminants, such as cattle, sheep and goats, has financial and welfare implications. The genetic interactions that lead to horn development are not known. Hornless, or polled, cattle occur naturally. The known causative DNA variants (Celtic, Friesian, Mongolian and Guarani) are in intergenic regions on bovine chromosome 1, but their functions are not known. It is thought that horns may be derived from cranial neural crest stem cells and the POLLED variants disrupt the migration or proliferation of these cells. Relaxin family peptide receptor 2 (RXFP2) is more highly expressed in developing horns in cattle compared to nearby skin and has been shown to play a role in horn development in sheep. However, the role of RXFP2 in horn formation is not understood. Histological analyses of cranial tissues from homozygous horned and polled cattle fetuses at day 58 of development was carried out to determine the differences in the structure of the horn bud region. Condensed cells were only observed in the horn bud mesenchyme of horned fetuses and could be the progenitor horn cells. The distribution of neural crest markers (SOX10 and NGFR) and RXFP2 between horned and polled tissues by immunohistochemistry was also analysed. However, SOX10 and NGFR were not detected in the condensed cells, and therefore, these cells are either not derived from the neural crest, or have differentiated and no longer express neural crest markers. SOX10 and NGFR were detected in the peripheral nerves, while RXFP2 was detected in peripheral nerves and in the horn bud epidermis. Previous research has shown that RXFP2 variants are associated with horn phenotypes in cattle an sheep. Therefore, the RXFP2 variants may affect the development of the epidermis or peripheral nerves in the horn bud.
Keywords
Headgear, polled, development, cattle, horn ontogenesis
Introduction
Horns are a type of headgear characteristic of ruminants, including cattle, goat and sheep. In some cattle production systems, the horns are removed from adults or prevented from developing in calves to improve the safety of stock handlers and other animals. However, these procedures have severe animal welfare implications (Sylvester et al., 2004; Mintline et al., 2013; Simon et al., 2022) and incur cost (Kling-Eveillard et al., 2015). So far, four dominant DNA variants (Celtic, Friesian, Mongolian and Guarani) on Bos taurus chromosome 1 have been found to cause hornlessness (polled) in cattle. As these variants are intergenic, the genes that directly affect bovine horn development are not known (reviewed by Aldersey et al., 2020).
In cattle, the adult horn consists of an inner core of bone surrounded by a keratin outer sheath. The horn is innervated by the corneal branch of the sensory trigeminal nerve (Buda et al., 2011). The process of horn development is not been well defined. However, the embryonic origin of horn tissues is likely to be from the ectoderm (skin), neuroectoderm (bone and nerves) and possibly mesoderm (bone).
The bovine horn bud is visible early in fetal development, at 60 days of gestation (Evans and Sack 1973), and histological studies have investigated horn bud structure from 70 days of gestation (Allais-Bonnet et al., 2013; Wiener et al., 2015; Schuster et al., 2020). The horn bud is usually compared to the frontal skin, which is in the cranial region corresponding to the frontal bone near the horn bud (Allais-Bonnet et al., 2013; Wiedemar et al., 2014; Wang et al., 2019). The epidermis at the horn bud is thicker than the epidermis of the frontal skin (Capitan et al., 2012; Allais-Bonnet et al., 2013; Wiener et al., 2015). Beneath the horn bud, thick nerve bundles are present from 115 days of gestation, whereas only normal nerve fibres are found in the frontal skin (Wiener et al., 2015). The horn bud also has delayed hair follicle development (Allais-Bonnet et al., 2013; Wiedemar et al., 2014; Wiener et al., 2015; Schuster et al., 2020), and no ossification is observed in the horn bud prior to birth (Wiener et al., 2015). The histological structure of the horn bud earlier than 70 days of fetal development has not been described.
The embryonic cell origin of the horn bud has not been investigated in bovine fetuses. It is currently thought that neural crest cells are responsible for horn ontogenesis (Wang et al., 2019; Aldersey et al., 2020). Cells expressing the neural crest markers SRY-box transcription factor 10 (SOX10) and neural growth factor receptor (NGFR, also known as p75NGFR or p75NTR) have been detected in ovine horn buds at day 90 of development (Wang et al., 2019; Rapizzi et al., 2020). SOX10 is a key regulator of neural crest cell development and maintains the cells in a stem cell state (Kim et al., 2003; Horikiri et al., 2017). NGFR is a member of the tumor necrosis receptor superfamily, and regulates neuronal processes including neuron cell survival, degradation and apoptosis (Goncharuk et al., 2020). To the best of our knowledge, SOX10 and NGFR expression have not been investigated in the bovine horn bud.
Few genes have been associated with horn development. The relaxin family peptide receptor 2 (RXFP2) gene has been implicated in horn development in sheep and cattle. In sheep, a variant in the 3`-UTR region of RXFP2 has been associated with horn status and shape (Wiedemar and Drögemüller 2015; Lühken et al., 2016). RXFP2 may also affect horn size and shape in ovine species (Kardos et al., 2015; Pan et al., 2018). In bovine and ovine fetuses at 70-90 days of development, expression of RXFP2 is higher in the horn bud compared to the frontal skin or the horn bud region in polled fetuses (Allais-Bonnet et al., 2013; Wiedemar et al., 2014; Wang et al., 2019; Wang et al., 2022).
Investigation of the histological structures in the early horn bud and localization of SOX10, NGFR and RXFP2 would provide further insight into bovine horn development. Therefore, this study characterized the bovine horn bud structure at 58 days of development by histomorphometric analysis, investigated the lineage of horn bud cells using the neural crest markers, SOX10 and NGFR, and determined the location of RXFP2 expression in the horn bud.
Results
Confirmation of fetal genotypes
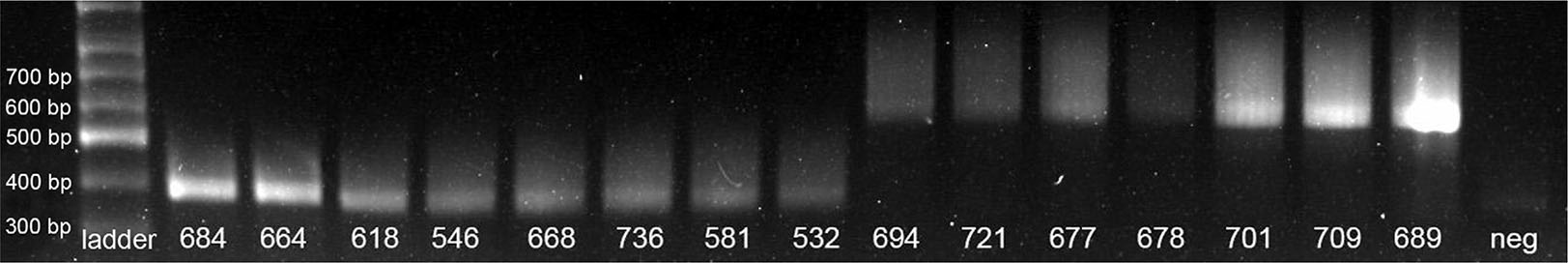
Five fetuses were collected for the pilot study and 11 more fetuses were collected for the main study (Table 1). Based on PCR genotyping, 15 fetuses aligned with their expected genotype (Fig. 1), but one fetus was heterozygous for the Celtic variant and was excluded from the study. In total, there were eight wild-type horned (pp) and seven homozygous polled (PcPc) fetuses.
Table 1
Fetuses collected for analysis, type of processing (haematoxylin and eosin [H&E] or immunohistochemistry [IHC]) and quality of histology samples
| Fetus ID | Trial | Age (days) | Genotype | Stain | Quality | |
|---|---|---|---|---|---|---|
| Horned (pp), polled (PP), or heterozygous (Pp) | HB | FS | ||||
| 618 | Main | 58 | Horned | H&E, IHC | 2 | 2 |
| 546 | Main | 58 | Horned | H&E, IHC | 3 | 1 |
| 736 | Main | 58 | Horned | H&E, IHC | 3 | 1 |
| 532 | Main | 58 | Horned | H&E, IHC | 3 | N/A |
| 668 | Main | 58 | Horned | H&E, IHC | 3 | 1 |
| 581 | Main | 58 | Horned | H&E, IHC | 3 | 1 |
| 694 | Pilot | 60 | Polled | H&E, IHC | 1 | 3 |
| 667 | Main | 58 | Polled | H&E, IHC | 3 | 1 |
| 701 | Main | 58 | Polled | H&E, IHC | 1 | 1 |
| 689 | Main | 58 | Polled | H&E, IHC | 1 | 1 |
| 709 | Main | 58 | Polled | H&E, IHC | 2 | 1 |
HB, horn bud; FS, frontal skin; 1, poor quality; 2, moderate quality; 3, good quality.
Macroscopic appearance and position of the horn bud
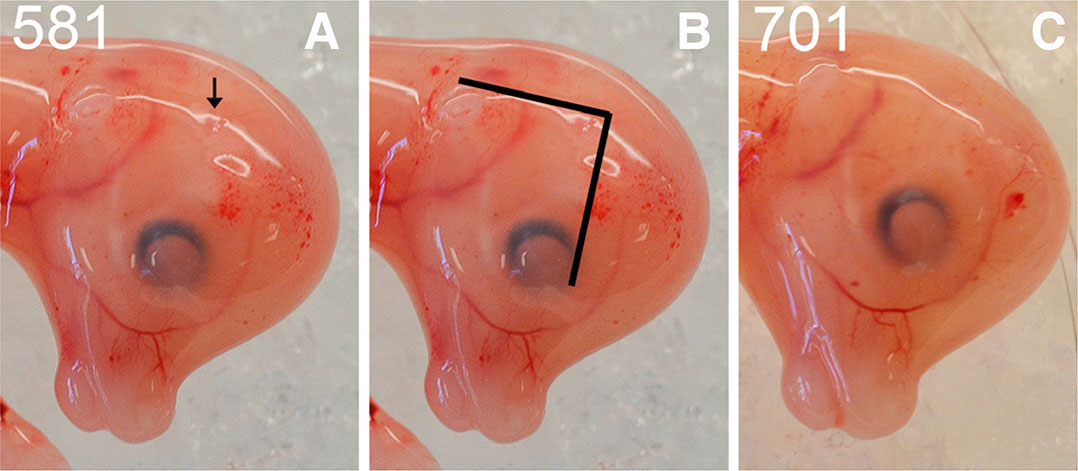
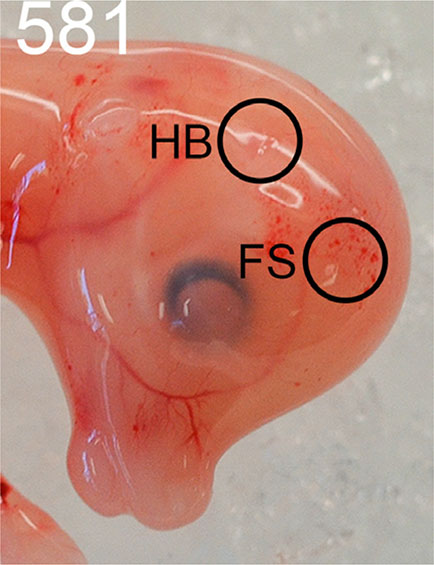
The horn bud in the PcPc fetuses was macroscopically visible as a slightly depressed ring of skin with a raised centre, which was absent in the polled pp fetuses. The position of the horn bud was at the intersection of perpendicular lines drawn from the eye and ear bud (Fig. 2). The skin was smooth in polled fetuses at the same position.
Fig. 2. Comparison of horned and polled fetuses at 58 days of development.
(A) Horned fetus example (#581) with an arrow indicating the horn bud. (B) The horn bud was positioned at the right angle of perpendicular lines drawn from the eye and ear bud. (C) Polled fetus example (#701) with no visible horn bud. At the horn bud region of polled fetuses, the skin was completely smooth.
Method testing
Histological analyses were carried out on six horn bud samples and five frontal skin samples collected from horned fetuses, and five horn bud regions and five frontal skin samples from polled fetuses. However, the frontal skin samples and the horn bud region samples from the polled fetuses were more fragile, and therefore were more easily damaged during processing. The samples that had structural damage to the tissue, such as missing epidermis, could not be used and only samples with good or moderate quality were measured. All six horn bud samples from horned fetuses were intact, whereas only one frontal skin sample from the horned fetuses had moderate quality (Table 1). Two polled horn bud samples and one polled frontal skin were undamaged and these were combined for the histomorphometric analysis (Table 1).
Horn bud tissue structures
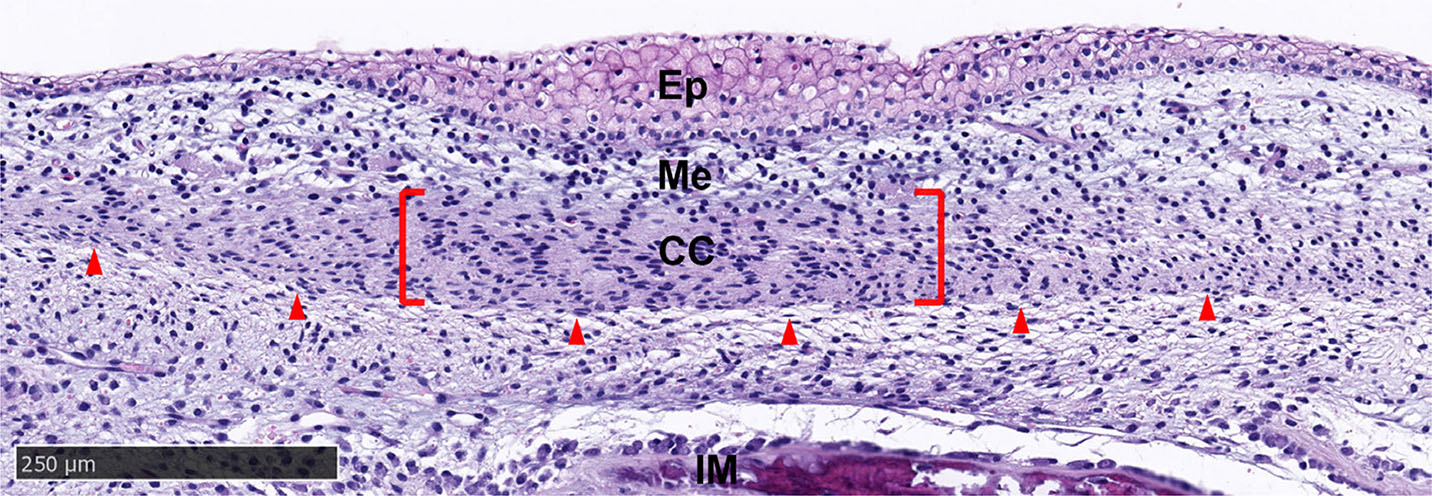
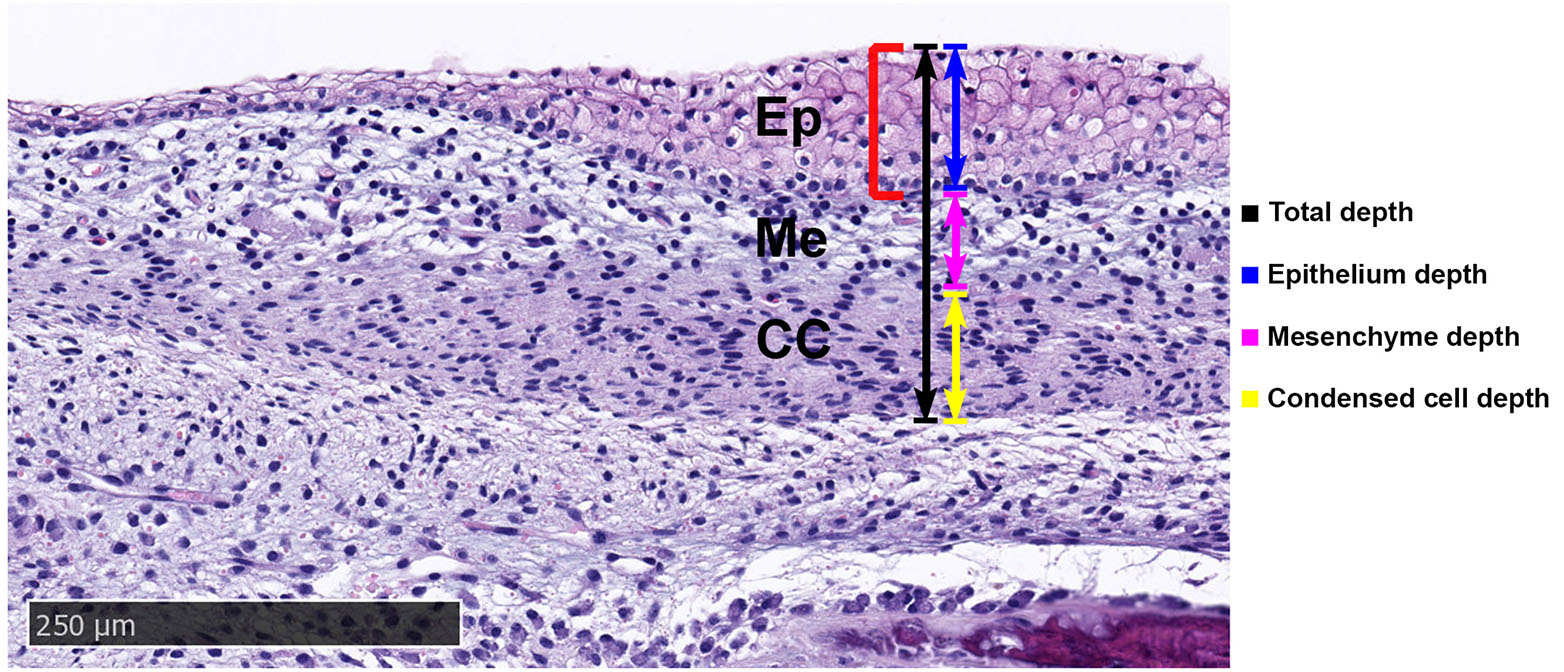
The horn bud at 58 days in the horned fetuses was characterised by a thickened epidermis of up to 10 cells at the centre, while surrounding tissue and frontal skin were only 1-2 cells thick. Five tissue layers were observed in the horn bud sections of the horned fetuses (Fig. 3). The outer epithelial layer was comprised of keratinocytes and was separated from the mesenchyme by the basal membrane. The mesenchyme layer had loosely packed cells that weakly stained with eosin and had round nuclei. The condensed cell layer had densely packed cells with oval nuclei and greater eosin staining than the mesenchyme (Fig. 3). An interstitial mesenchyme layer of 1-3 cells with oval/elongated nuclei demarcated the lower edge of the condensed cell layer. Below the interstitial mesenchyme, ossification was observed as samples were taken at the frontal bone region.
Fig. 3. Tissue layers in the horn bud section from horned fetus #668 stained with haematoxylin and eosin.
Condensed cells are located below the thickened epithelium of the horn bud (red bracket). A thin layer of interstitial mesenchyme (red arrows) separates the condensed cells (CC) and developing skull vault (IM). Ep, epithelium; IM, intramembranous ossification; Me, mesenchyme. Magnification, 10x.
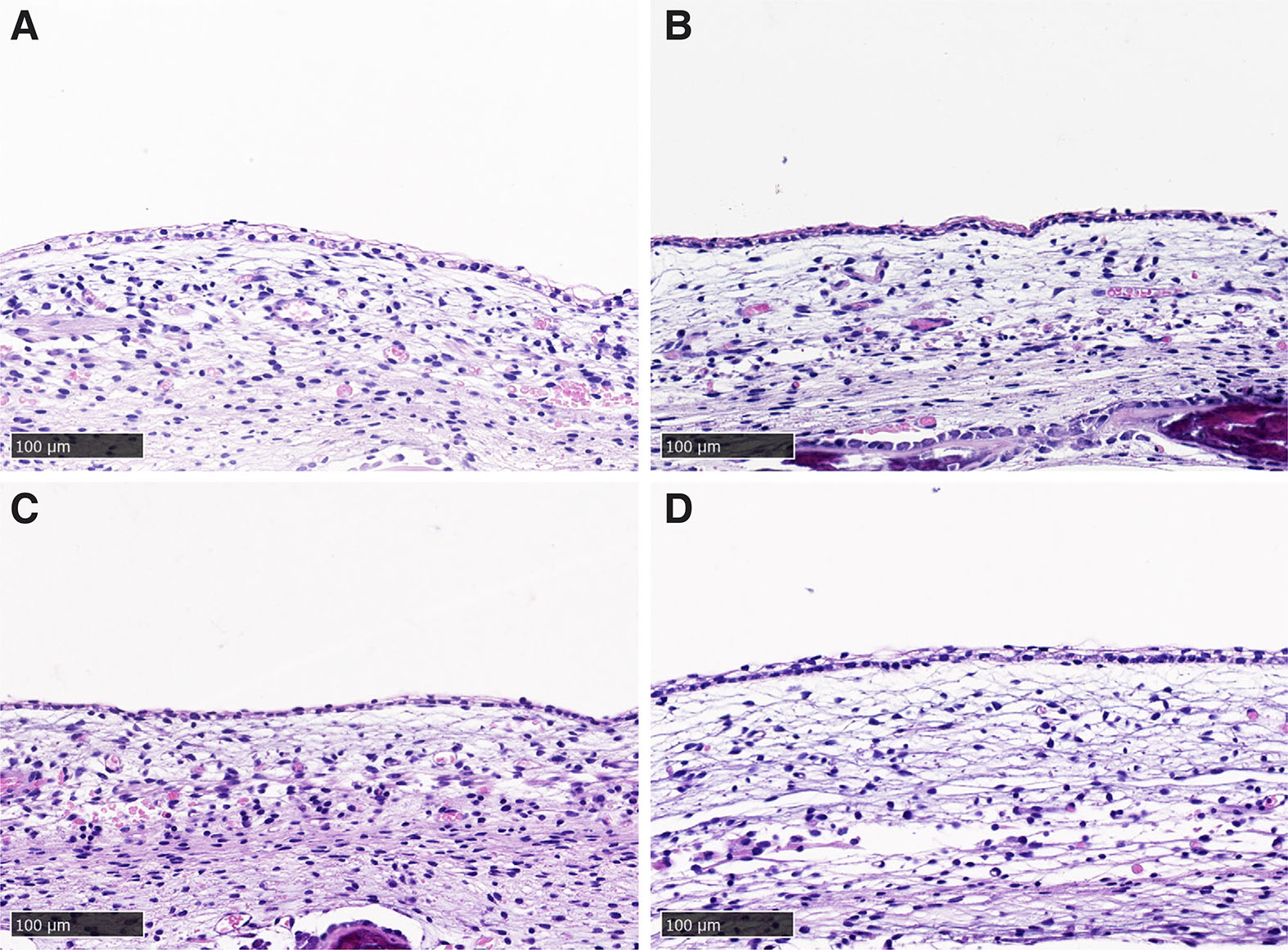
The tissue outside the horn bud of horned fetuses and tissue from polled fetuses had an epithelial cell depth of 1-2 cells (Fig. 4). The same tissue layers were present as within the horn bud, but the condensed cell depth was less apparent or absent. The tissues had a similar appearance between the outer horn bud and frontal skin from horned fetuses (Fig. 4 A,B).
Comparison of the inner horn bud, outer horn bud and polled regions
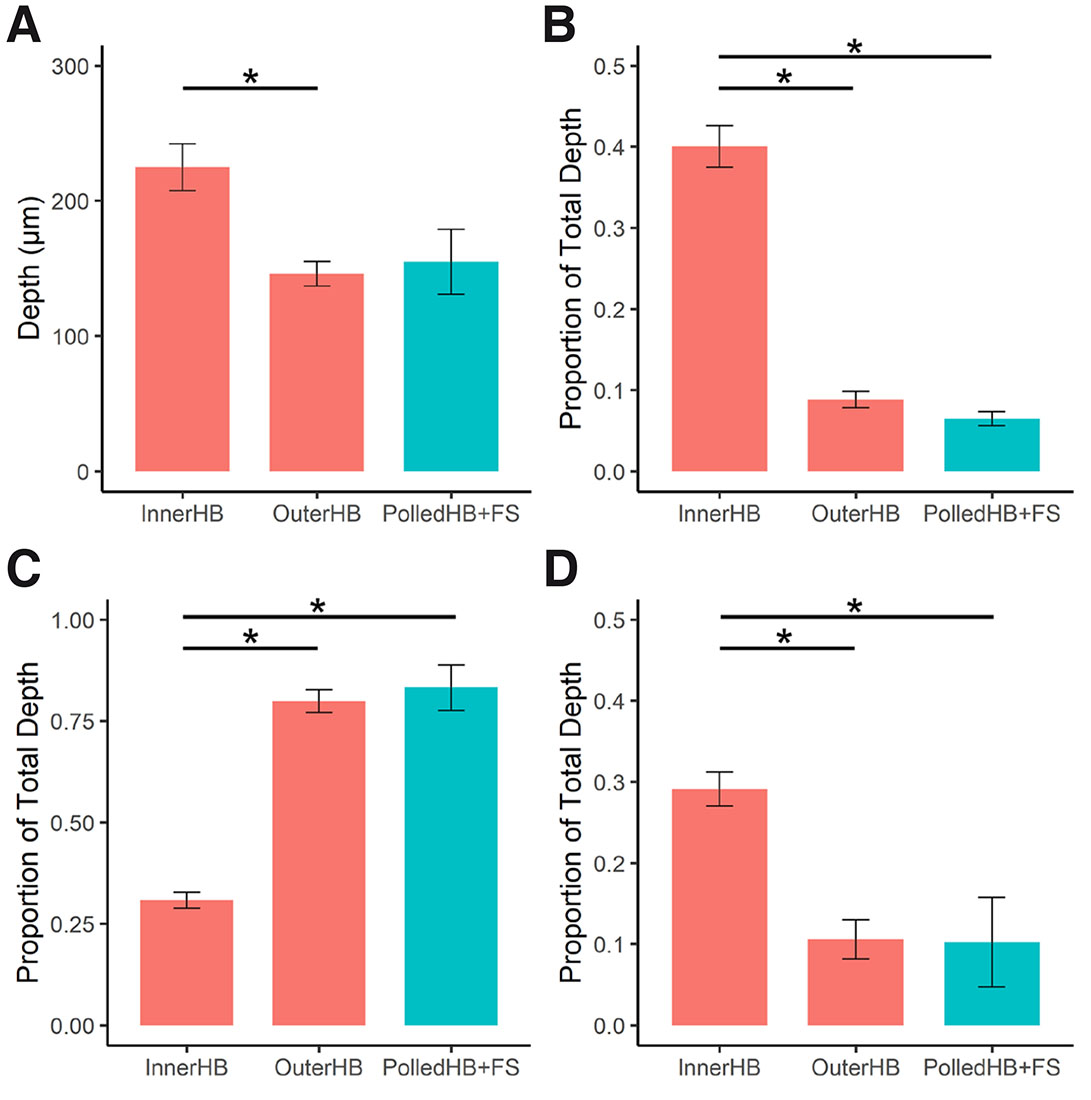
The measurements from the inner horn bud (InnerHB; Fig. S1) and outer horn bud (OuterHB; Fig. S2) regions were compared with the polled horn bud regions and frontal skin samples combined (PolledHB+FS) (Table 2). The measurements from the InnerHB were significantly different from the OuterHB and from the PolledHB+FS samples (Fig. 5), indicating that the tissue at these sites was structurally different. Overall, the InnerHB had a greater total depth, epithelium depth, and condensed cell depth, while the mesenchyme depth was greater in the OuterHB and PolledHB+FS. The horn bud region of polled fetuses and frontal skin sections were indistinguishable histologically.
Fig. 5. Cell depth measurements.
Comparison of (A) total depth, (B) epithelium depth, (C) mesenchyme depth and (D) condensed cell depth, between the inner horn bud (InnerHB), outer horn bud (OuterHB) and tissue from the polled horn bud region and frontal skin (PolledHB+FS). Horned fetuses, red; polled fetuses, blue. Epithelium depth, mesenchyme depth and condensed cell depth are reported as proportions of the total depth. Columns and error bars represent the mean proportion and standard error. Paired and unpaired Wilcoxon rank sum tests were conducted to compare medians between groups. * p < 0.05.
Table 2
Descriptive statistics of measurements obtained from H&E horn bud (HB) and frontal skin (FS) samples
| n | Total (µm) | Epithelium (µm) | Mesenchyme (µm) | Condensed cell (µm) | |||||
| Median | SD | Median | SD | Median | SD | Median | SD | ||
| InnerHB | 6 | 228.2 | 42.5 | 83.3 | 19.5 | 71.2 | 20.1 | 65.3 | 16.9 |
| OuterHB | 6 | 151.3 | 22.2 | 11.9 | 4.4 | 114.1 | 20.6 | 14.5 | 9.8 |
| PolledHB+FS | 3 | 163.0 | 41.5 | 9.0 | 3.1 | 141.8 | 47.4 | 12.7 | 10.1 |
Localisation of SOX10 and NGFR
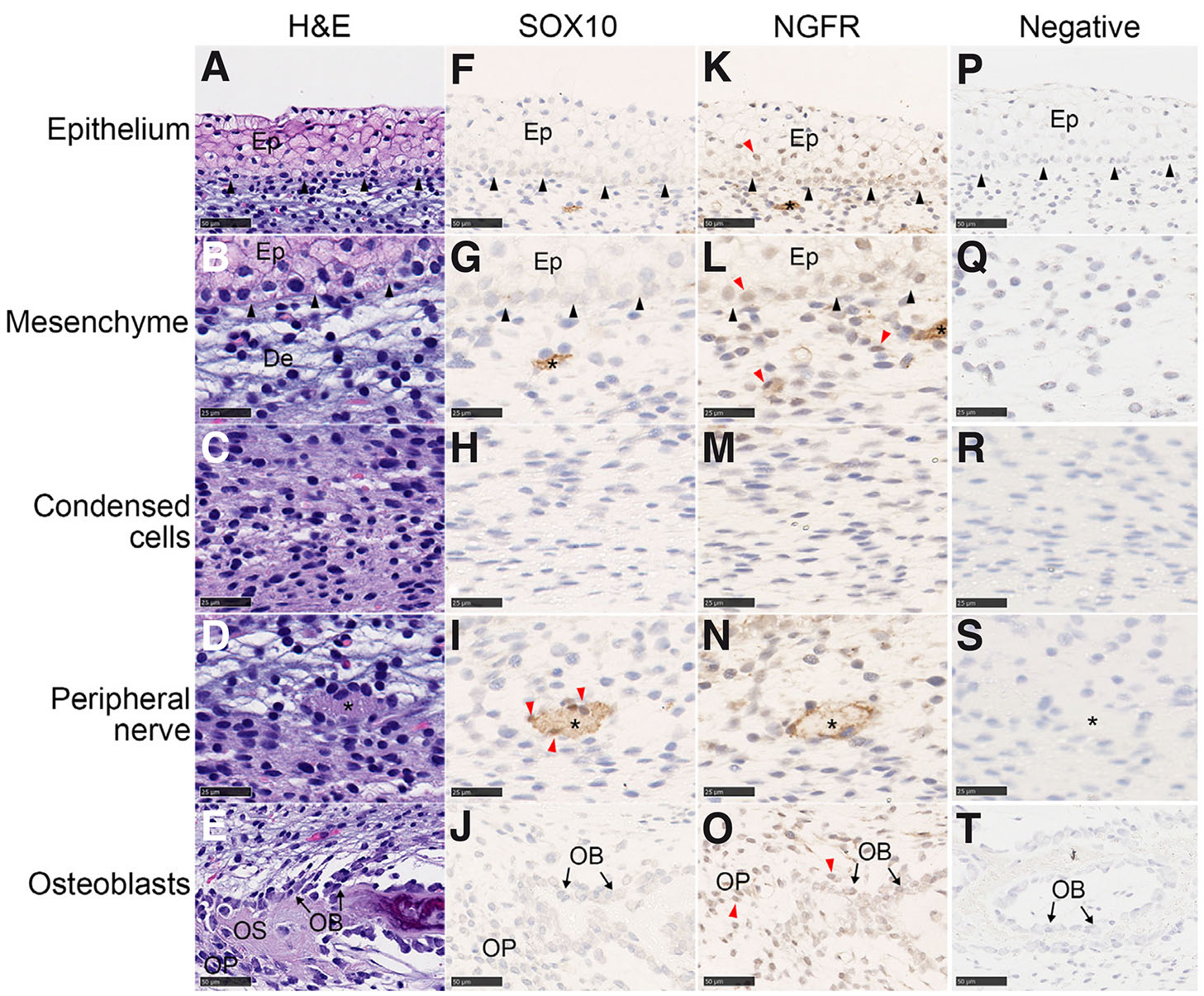
Horned and polled samples were stained with antibodies for the neural crest markers, SOX10 and NGFR, to determine if cells in the sections were derived from neural crest cells. In the horn bud of horned fetuses, SOX10 and NGFR expression was detected in structures that possibly develop into peripheral nerves (Fig. 6; Fig. S3 A-B). Cells associated with peripheral nerves, glial cells, were also positively stained by SOX10. Growth of peripheral nerves into the epidermis was observed for five out of six horned HB samples (Fig. S3 C,D). Neither SOX10 nor NGFR were detected in the condensed cell layer (Fig. 6 H,M). In contrast to SOX10, NGFR was widely expressed in the horn bud. In addition to expression in peripheral nerves (Fig. 6N), NGFR was expressed in cells in the epidermis, mesenchyme and developing cranial vault in the horn bud samples (Fig. 6 K-L, O).
Fig. 6. SOX10 and NGFR antibody staining of cells in the horn bud of horned fetuses.
SOX10 and NGFR are present in the peripheral nerves. SOX10 was also found in some glial cells. NGFR was found in the epithelium, some mesenchymal cells and the developing cranial vault. The horn bud was stained with haematoxylin and eosin (A-E), SOX10 antibody (F-J) and NGFR antibody (K-O) and counterstained with haematoxylin. Negative controls (P-T) were stained without the primary antibody and counterstained with haematoxylin. Positively staining nuclei were observed for SOX10 (glial cells) and NGFR (epithelium, mesenchyme and osteoblasts) (red arrows). Nerve tissue is indicated with an asterisk (*) and basal epithelium by black arrows. Ep, epithelium; OB, osteoblast; OP, osteoprogenitor; OS, osteoid. Magnifications: 40x (A, F, K, E, J and O; scale bar, 25 µm) and 80x (B-D, G-I, L-N; scale bar, 50 µm).
The SOX10 and NGFR antibody staining of the polled tissues reflected the same pattern as observed in the horn bud of the horned fetuses (Fig. S4). SOX10 was detected in the glial cells and developing nerves, although the nerves were smaller than those in the horn bud (Fig. S4C). Individual cells that were not near the nerves, but still expressing SOX10, were apparent in some sections (Fig. S4A). This may have occurred because the section included nuclei but not the whole nerve. NGFR was detected in the developing nerves, mesenchymal cells, epithelium cells and the developing cranial vault (Fig. S4 B-D). No nerves were detected within the epithelium. The polled frontal skin samples were similar to the polled horn bud region samples.
Localisation of RXFP2
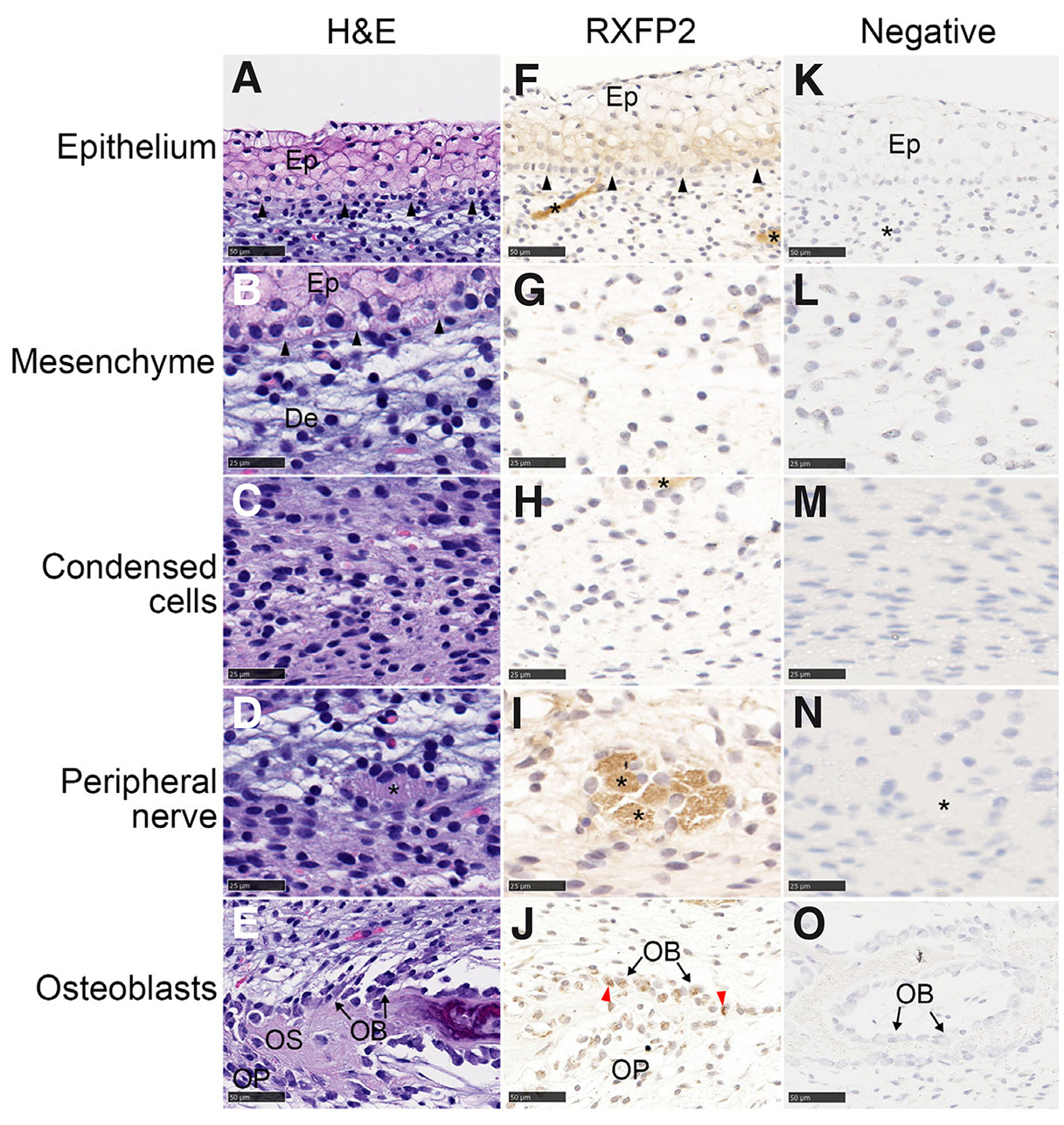
Using RXFP2 antibody staining of cells in the horn bud of horned fetuses, RXFP2 was detected in the epithelium, developing peripheral nerves and osteoblasts (Fig. 7). There was also low expression in the endothelial cells of blood vessels. In the polled tissues, RXFP2 staining was observed in nerve tissues, osteoblasts and osteoprogenitor cells (Fig. S5). However, RXFP2 was not seen in the polled epithelium.
Fig. 7. RXFP2 antibody staining of cells in the horn bud of horned fetuses.
RXFP2 is present in the epithelium and developing peripheral nerves. The horn bud was stained with haematoxylin and eosin (A-E) and RXFP2 antibody and counterstained with haematoxylin (F-J). Negative controls (K-O) were stained without the primary antibody and counterstained with haematoxylin. Nerve tissue is indicated with an asterisk (*). Ep, epithelium; OB, osteoblasts; OP, osteoprogenitor cells; OS, osteoid; red arrows, positively stained nuclei; black arrows, basal membrane. Magnification, 40x (A, E-F, J-K, O; scale bar, 50 µm) and 80x (B-D, G-I, l-N; scale bar, 25 µm).
Nerve tissue area measurements
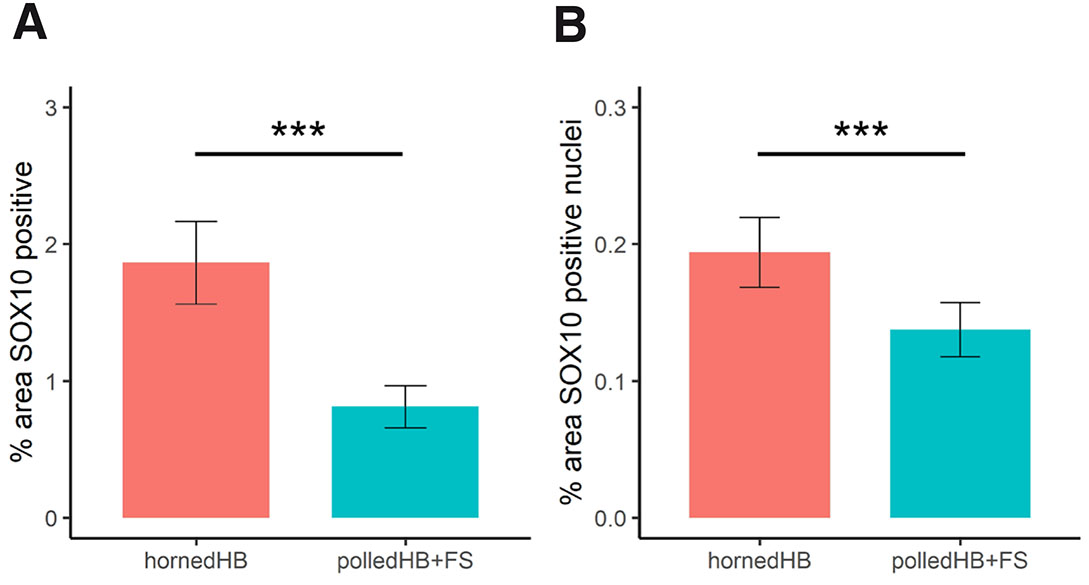
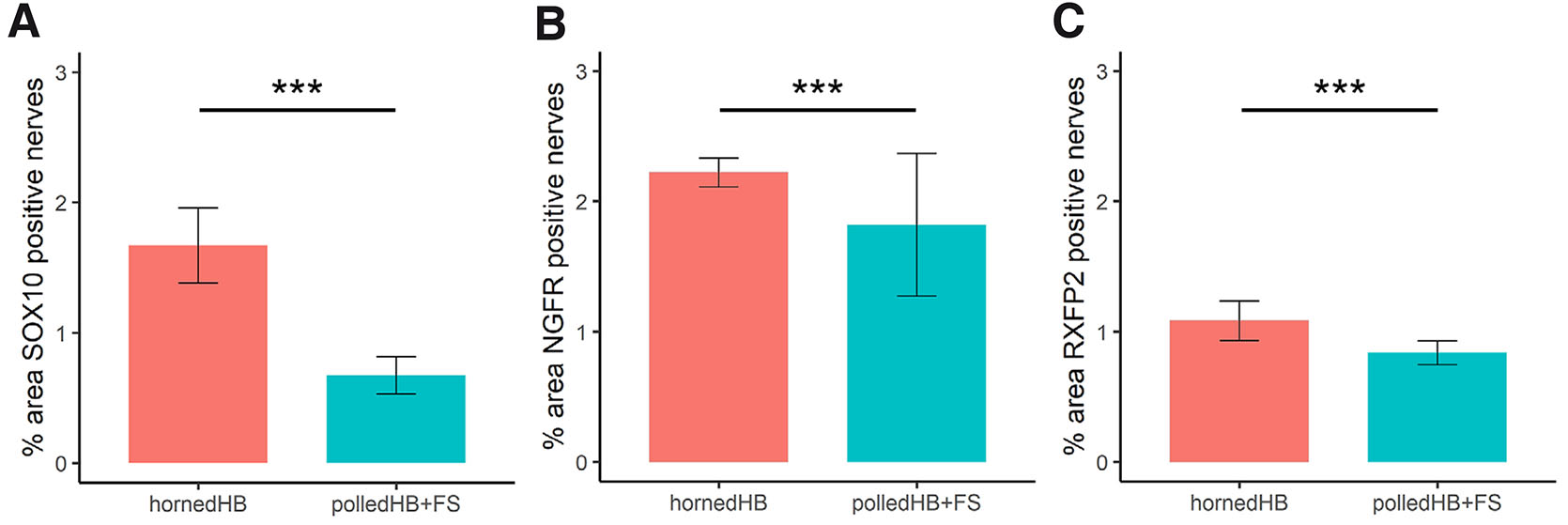
To test whether there was more nerve tissue in the horn bud, machine learning was used to segment the section images into nerves, nuclei and background, and then the areas were measured in pixels. Additionally, for the SOX10 sections, positive and negative staining nuclei were differentiated. Nerves identified by positive staining were compared between horned and polled tissues (Table 3). In total for SOX10 (cell nuclei and nerves), there was 1.67 times more pixels with positive staining in horn bud tissues compared to polled tissues (Fig. 8A). In addition, there was 1.67 times more pixels labelled as positively stained cells for SOX10 in the horn bud of horned fetuses than in polled HB and FS (Fig. 8B). Based on the SOX10 staining, there were 3.15 times (odds ratio) more pixels labelled as nerves in the horn bud from horned fetuses compared to the corresponding region in the polled tissues (HB+FS) (Fig. 9A). These results indicate that there is significantly more nerve tissue developing in the horn bud of horned fetuses compared to horn bud regions and frontal skin of polled fetuses.
Fig. 8. Positive staining of SOX10 in the horn bud of horned fetuses (HB, n = 5) and polled tissues (HB+FS; n = 3).
(A) Total positive staining (positive nuclei and nerves) and (B) positively stained nuclei presented as a percentage of the total tissue area. Columns and error bars represent the mean proportion and standard error. *** p < 0.001.
Fig. 9. Area of nerves postively stained by anti-SOX10, anti-NGFR and anti-RXFP2.
Nerves stained positively by (A) SOX10, (B) NGFR and (C) RXFP2 in the horn bud of horned fetuses (HB, n = 5) and polled tissues (HB+FS; n = 3) presented as a percentage of the total tissue area. Columns and error bars represent the mean proportion and standard error. *** p < 0.001.
Table 3
Comparison of positively stained areas for SOX10, NFGR and RXFP2 between horned horn bud (n = 6) and polled horn bud + frontal skin (n = 3) using the Fisher’s exact test
| Stain | Target Area | P-value | Confidence interval |
|---|---|---|---|
| SOX10 | Positive cell nuclei + Positive nerves | < 2.2e-16*** | 2.87 – 2.92 |
| Positive cell nuclei | < 2.2e-16*** | 1.64 – 1.71 | |
| Positive nerves | < 2.2e-16*** | 3.12 – 3.18 | |
| NGFR | Positive nerves | < 2.2e-16*** | 1.36 – 1.38 |
| RXFP2 | Positive nerves | < 2.2e-16*** | 3.12 – 3.18 |
*** p < 0.001
The nerve tissue was also measured in the NGFR and RXFP2 stained sections (Table 3). For the NGFR stained sections, there were 1.37 times more pixels labelled as nerve tissue in the horn bud of horned fetuses compared to polled HB+FS (Fig. 9B). For the RXFP2 stained sections, there was 1.25 times more positive staining in the horn bud compared to the polled tissues (Polled HB+FS; Fig. 9C). These results are consistent with the results from staining with the SOX10 antibody and again indicated that there was more nerve tissue in the horn bud of horned fetuses compared to the polled tissues.
Discussion
In this study, the bovine horn bud structure was characterised at 58 days of fetal development by histomorphometric analysis. Obtaining high quality fetal samples for staining proved difficult, particularly for the polled samples which were very fragile. Nevertheless, more samples were analysed than in previous histological studies of horn development wherein only a single fetus was sampled per time-point and genotype (Allais-Bonnet et al., 2013; Wiener et al., 2015).
The analysis revealed a thickened epidermis and layer of condensed cells below the mesenchyme in the horn bud that was not present in the comparable regions from polled fetuses (Fig. 3). This is consistent with previous studies that found that the epithelium layer was thicker in the horn bud compared to frontal skin in horned fetuses at 70-90 days of development (Allais-Bonnet et al., 2013; Wiener et al., 2015; Wang et al., 2022). A layer of condensed cells was observed in horn bud of horned fetuses, which was significantly reduced or absent outside the horn bud and in polled tissues. Condensed cells below the mesenchyme in the horn bud of horned fetuses were not reported in a previous histological study of fetal horn bud at 70 days of development (Wiener et al., 2015). The horn bud at 70 days had a similar structure to the horn bud from fetuses at 58 days in the current study, but the condensed cell layer was labelled ‘dermis’. However, the cell types in ‘dermis’ of the horn bud and frontal skin are dissimilar, as the frontal skin does not have the condensed cells. The histology sections from the horn bud and frontal skin at 70 days support our findings (Wiener et al., 2015).
The identity of the condensed cells is not known, but they could be horn progenitor cells that have aggregated to undergo further differentiation. Aggregating cells often initiate organogenesis, including the development of bone, hair and teeth (Fuchs 2007; Li et al., 2016; Puthiyaveetil et al., 2016; Salhotra et al., 2020). In the case of hair and teeth, interactions between the epithelium and the condensed cells are an essential part of organogenesis (Fuchs 2007; Puthiyaveetil et al., 2016). As teeth develop, the mechanical compression caused by the expanding epithelium leads to the aggregation of underlying mesenchymal cells (Svandova et al., 2020).
Cells important for horn development are thought to be derived from the cranial neural crest. However, the neural crest markers, SOX10 and NGFR, were not detected in the condensed cell layer. It is possible that these cells may have differentiated, and no longer express these neural crest markers, or that the horn bud is not derived from the neural crest. Alternatively, the condensed cells may not be the horn progenitors. Therefore, investigation of neural crest markers at earlier time points than day 58 or the use of different markers is necessary to determine cellular origin of the horn progenitor cells.
The SOX10 staining pattern in the bovine horn bud was different from the expression observed in the horn bud of ovine fetuses (Wang et al., 2019). In the present bovine study, only a few cells with strong SOX10 staining were detected. In contrast, the ovine horn bud at approximately 90 days of development had positively stained cells in the epithelium and the mesenchyme (Wang et al., 2019). The difference in staining observed may be due to the different developmental stage of the fetuses studied, as the pregnancy lengths in cattle and sheep are not the same (~9.5 months in cattle and ~5 months in sheep). Interestingly, the NGFR staining pattern in the present study was consistent with the pattern seen in ovine horn bud.
SOX10 and NGFR were detected in structures presumed to be immature peripheral nerves in the fetal horn bud. NGFR, which binds to neurotrophins, is involved in neuronal survival, neurogenesis, neurite outgrowth and apoptosis (Lu et al., 2005). Cells associated with the peripheral nerves expressed SOX10, which is consistent with glial cells, such as pro-myelinating Schwann cells. The innervation in the horn bud of horned fetuses and the horn bud region and frontal skin from polled fetuses was significantly different. More peripheral nerve tissue was detected in the horn bud than in the polled tissues. Wiener et al., (2015) observed ‘thick nerve bundles’ in the horn bud of fetuses at 115 and 140 days of development and comparatively smaller nerves in polled fetuses that were the same age. These nerves are likely to develop into the corneal branch of the sensory trigeminal nerve (Buda et al., 2011).
RXFP2 was detected in the epithelium, developing peripheral nerves and osteoblasts in the horn bud samples, but not in the epithelium of polled samples. RXFP2 is differentially expressed between horned and polled bovine and ovine fetuses at 90 days (Allais-Bonnet et al., 2013; Wiedemar et al., 2014). RXFP2 has been also associated with horn status and shape in sheep (Kardos et al., 2015; Wiedemar and Drögemüller 2015; Duijvesteijn et al., 2018; He et al., 2020), and linked to scurs in sheep (Johnston et al., 2011) and cattle (Wang and Gill 2021). The evolutionary loss of antlers in musk deer and Chinese water deer is attributed to nonsense and missense mutations in RXFP2 (Wang et al., 2019). The observation in the present study that RXFP2 is present in developing peripheral nerve and horn bud epithelium suggests the development of these tissues may be affected by RXFP2, and may potentially lead to phenotypic differences in horn status.
Conclusion
Macroscopic and microscopic differences between horned and polled tissues of the horn bud region were identified at day 58 of bovine fetal development. In addition to a thicker epidermis, which has been observed in earlier studies, an aggregation of cells was seen in the horn bud mesenchyme that may represent horn progenitor cells. At this stage of fetal development, these presumptive horn progenitor cells did not express the neural crest markers SOX10 and NGFR, suggesting that cranial neural crest cells do not contribute to horn ontogenesis, or that these cells have already differentiated by day 58 of bovine fetal development. At day 58, developing peripheral nerves were positive for both NGFR and SOX10, but glial cells, which are potentially pro-myelinating Schwann cells, were only positive for SOX10. RXFP2 was found in the horn bud epithelium and peripheral nerves, suggesting a role for RXFP2 in the formation these structures. Previous studies have suggested that RXFP2 is associated with horn development in cattle and sheep. In this study, RXFP2 was localised to the epithelium and peripheral nerves, suggesting that these tissues could be involved in horn development. Future research should further characterise the location of RXFP2 in the bovine and ovine horn bud region of horned, scurred and polled fetuses at different ages.
Materials and Methods
Animals
In total, 12 horned and 12 polled Hereford heifers were used in this study, which was approved by the University of Adelaide Animal Ethics Committee (Project Approval No. S-2018-105).
Determination of appropriate fetal age for tissue collection
A preliminary trial was conducted to determine the earliest age for fetal collection that would enable dissection of the horn bud. Six homozygous horned (p/p) and six homozygous polled (PC/PC) Hereford heifers from a mixed phenotype herd were synchronised for artificial insemination. The horned heifers were artificially inseminated with semen from a horned bull (p/p) and polled heifers inseminated with semen from a polled bull (PC/PC).
One horned and one polled fetus was surgically removed by a veterinarian at 58 days, and one horned and two polled fetuses were collected at 60 days. The fetuses were recovered by left sided, oblique laparotomy in the caudal paralumbar fossa, under local anaesthesia (distal paravertebral block followed by inverted L in the left flank). The fetuses were placed in sterile containers and immediately transported on ice to the laboratory. The horn bud was visible at 58 days. Therefore, the additional fetuses were subsequently collected at 58 days.
Collection of 58 day old fetuses for analysis
Horned and polled heifers (n = 19) were allocated into two synchronization groups. Groups were offset by two days to allow time for all fetuses to be collected. The two groups had five and six pregnant heifers. At 58 days of gestation, the fetuses were surgically removed, placed in sterile containers, and immediately transported on ice to the laboratory.
The fetal heads were dissected in half, rostral-caudal. One half of each head was preserved in formalin fixative solution. After fixation for more than 48 hours at room temperature, the samples were dehydrated in a graded ethanol series at 25% and 50% for 24 hours each and then 75% ethanol. The brain tissue was removed from the cranial cavity and a 4 mm biopsy punch was used to sample the horn bud (HB) region and frontal skin (FS) region (Fig. 10). The other half of the head was preserved for additional analysis not described here.
Genotyping for Celtic variant by PCR
DNA was extracted from adult ear notches and fetal skin samples using the Qiagen DNeasy Blood and Tissue kit. Primers encompassing the Celtic variant location were used for PCR amplification (btHP-F1: 5’-GAAGGCGGCACTATCTTGATGGAA; btHP-R1: 5’-GGCAGAGATGTTGGTCTTGGGTGT) (Carlson et al., 2016). The PCR assay was conducted using the KAPA Taq ReadyMix PCR Kit (Kapa Biosystems, Inc.). An initial melt was at 95°C, before denaturation (95°C for 20s), annealing (62°C for 20s) and extension (72°C for 20s) for 34 cycles, followed by a 1 min extension at 72°C. A 389 bp product was amplified from the horned sequence and a 591 bp product was amplified from the Celtic polled sequence.
Tissue processing for histochemical and immunochemical analyses
Tissue processing was carried out by the Histology Services in the Adelaide Medical School at the University of Adelaide. The ethanol-dehydrated samples were cleared with xylene before embedding in paraffin with the tissue processor Tissue-Tek VIP 6 AI (Sakura). Samples were sectioned at 4 μm thickness using a microtome (RM2235, Leica) as this was the thickness used by Wiener et al., (2015). Four sections were placed on each slide, and every other slide was stained with haematoxylin and eosin (H&E). The unstained slides were retained for immunohistochemistry (IHC). Temporal lobe samples from a calf that died from dystocia and bull testicular tissue samples obtained from an abattoir were sectioned at 5 μm thickness and used as positive controls for IHC antibodies.
Haematoxylin and eosin (H&E) staining
The slides were dipped in xylene, absolute ethanol, and 70% ethanol before staining with H&E. To stain, the slides were dipped in Harris Haematoxylin (Dako, CS709) for 1 min, rinsed with tap water for 1 min, dipped in Bluing Buffer (Dako, CS702), rinsed with tap water for 1 min, incubated in 70% ethanol, then incubated in Eosin Y with Phloxine B (Dako, CS710) for 4.5 min. The slides were washed with absolute ethanol, xylene and Histo-Clear, and then allowed to dry before applying a coverslip.
Immunohistochemistry (IHC)
For the immunohistochemistry, the slides were dipped in xylene, and rehydrated by serial washes in 100%, 95% and 75% ethanol solutions and then placed in distilled water.
For antigen retrieval, the slides were heated to boiling point in a microwave oven in EDTA buffer (0.5) for 10 min, and then cooled. Once the slides reached room temperature (RT), they were incubated in 20 mM Tris Buffered Saline with 0.1% sodium azide (TBS-azide) for 10 min at RT. Endogenous peroxidase activity was blocked by incubating sections with a 1% H2O2-50% methanol solution for 10 min at room temperature, followed by a wash in TBS-azide for 10 min. Non-specific binding sites were blocked with incubation for 60 min in a humidity chamber in 20% normal horse serum in TBS-azide solution. The slides were dried around the sections using Kimwipes (KimTech Science) and the primary antibody was applied. Anti-SOX10 (Santa Cruz Biotechnology, Inc., sc-365692), anti-NGFR (Santa Cruz Biotechnology, Inc., sc-271708) and anti-RXFP2 (Santa Cruz Biotechnology, Inc., sc-374293) antibody dilutions were prepared using 1% normal horse serum in TBS-azide (Table 4). One section was incubated with only the antibody diluent solution (no antibody) as a negative control. The sections were incubated overnight in a humidity chamber at room temperature.
Table 4
Primary antibody dilutions used for immunohistochemistry
| Antibody | Dilution | Secondary antibody dilution |
|---|---|---|
| SOX10 | 1:200 | 1:200 |
| NGFR (P75NGFR) | 1:300, 1:500 | 1:200 |
| RXFP2 | 1:20 | 1:100 |
After incubation with the primary antibody, the slides were washed in TBS-azide, dried, and incubated with a biotin goat anti-mouse IgG Cross-Adsorbed Secondary Antibody (ThermoFisher Scientific, #62-6540 diluted in antibody diluent solution) for 90 min at room temperature in a humidity chamber. The slides were then washed in TBS-azide and incubated with avidin and biotin solution (VECTASTAIN ABC Kit, Vector Laboratories, Inc.) for 60 min at RT. The slides were washed in TBS and incubated for 3-5 min with a 3,3'-diaminobenzidine substrate solution which was prepared by dissolving a SIGMAFASTTM 3,3’-Diamino-benzidine tablet (SIGMA-ALDRICH Co.; product code: 1002771577) in 5 ml distilled water and 3.5 µl hydrogen peroxide. The reaction was stopped by rinsing the slides in TBS-azide. The slides were counterstained with haematoxylin (Mayer’s Hematoxylin Solution, product number: MHS1, Sigma Aldrich) and once dry, coverslips were applied.
Antibody validation
Specificity of the primary antibodies was tested by staining bovine tissues known to express the proteins of interest. The Sox10 antibody was assessed by staining bovine temporal lobe and the p75ngfr and Rxfp2 antibodies were assessed using bovine testicular tissue sections. Positive staining was seen in the expected regions (Fig. S6-S8). Fetal sections incubated without the primary antibody did not show positive staining.
Imaging
Slides were scanned using a NanoZoomer 2.0-HT slide scanner (Hamamatsu; model: C9600-01). The high-resolution scans (stored as.ndpi files) were viewed using NDP.view2 software (Hamamatsu). Images of the H&E stained slides were exported at 20x magnification as JPEG files (300 dpi). The IHC stained slides were exported at 5x magnification as JPEG images (2000 dpi).
Histomorphometrics
The number of epithelium cells was counted at the thickest part of the epithelium in the horned and polled samples (Fig. 11). Sections were grouped based on their genotype (horned or polled) and their location in the horn bud tissue (outside or center) using the depth of the epithelial cells as an indicator of position. For the horn bud tissues, sections with a depth of 1-2 epithelial cell layers were considered to be outside of the horn bud (OuterHB, n = 6; Fig. S1), whereas sections with a depth > 7 epithelium cells were considered to be at the center of the horn bud (InnerHB, n = 6; Fig. S2). Due to low sample numbers, the horn bud region of polled fetuses and frontal skin samples were grouped together (Polled HB+FS, n = 3).
Measurements were taken in micrometres (µm) on the H&E sections using FIJI ImageJ software (Schindelin et al., 2012). Four tissue measurements were taken: total depth, epithelial depth, mesenchyme depth and condensed cell depth (Fig. 12). The first section from every H&E stained slide was measured, which equated to every eighth section, giving ~28 µm between each measurement.
Image segmentation analysis
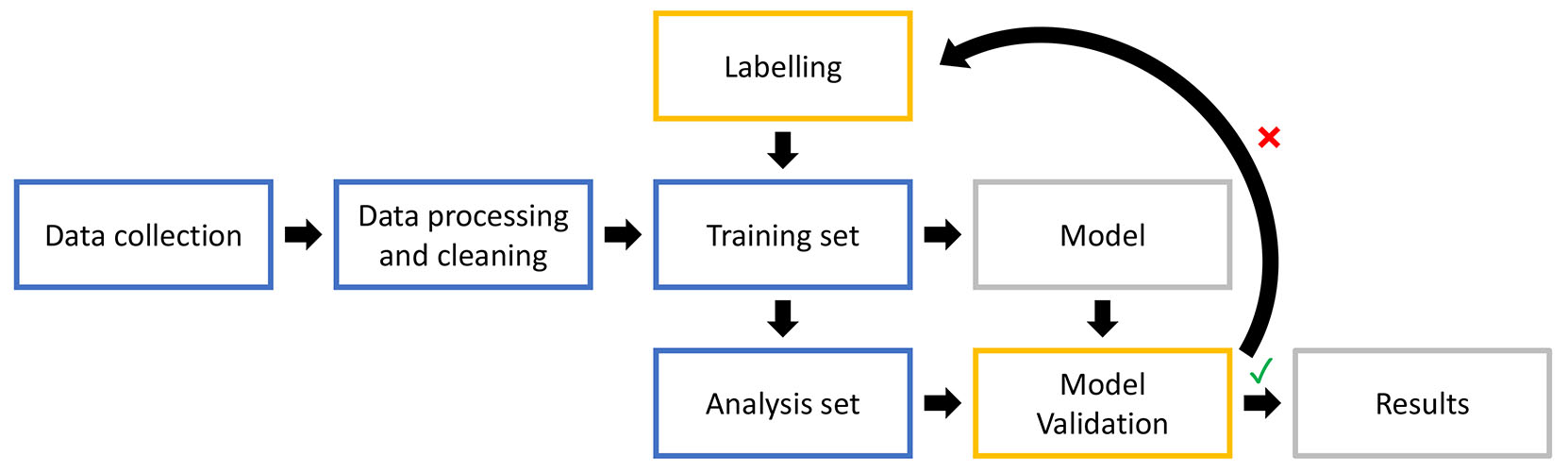
Images of IHC slides were manually processed in Photoshop (Adobe Creative Cloud 2018) to select the dermal tissue for image segmentation analysis. The epithelium and developing cranial bone were excluded from the analysis. The.jpeg images were converted to.czias files using ZEN 3.4 (Zen lite; Carl Zeiss Microscopy GmbH).
The IHC images were segmented using a supervised machine learning approach in Intellesis (ZEISS) (Fig. 12). Prior to training, the images were grouped based on staining strength (medium, light or dark). Images were removed from analysis if the section was damaged or if the stain was too light or too dark. For each tissue sample, three stained sections per antibody were randomly selected and combined for analysis (‘analysis set’). Medium stained sections were prioritised over light and dark sections in order to maximise the number of sections that were analysed with the same model. Using the approach of Nesbit et al., (2021), six to ten sections were allocated to the ‘training set’ to create the image segmentation models. The model was then tested on the analysis set and the accuracy was scored as good or poor. Training and testing were repeated until most sections had good segmentation. Some sections consistently did not fit the model, and therefore, separate models were trained to analyse these sections.
To train the model, images were labelled for the following regions: “background”, “negative nuclei” and “positively stained regions” (Fig. 13 A,B). For the anti-SOX10 labelled sections, the “positively stained nuclei” were detected as an additional region (Fig. 13 C,D). Minimum areas were set at 200 pixels for “positively stained regions” and “nuclei”. For anti-SOX10 stained sections, minimum areas were set to 250 pixels for “positively stained nuclei”, 300 pixels “positively stained regions” and 150 pixels for “negative nuclei”.
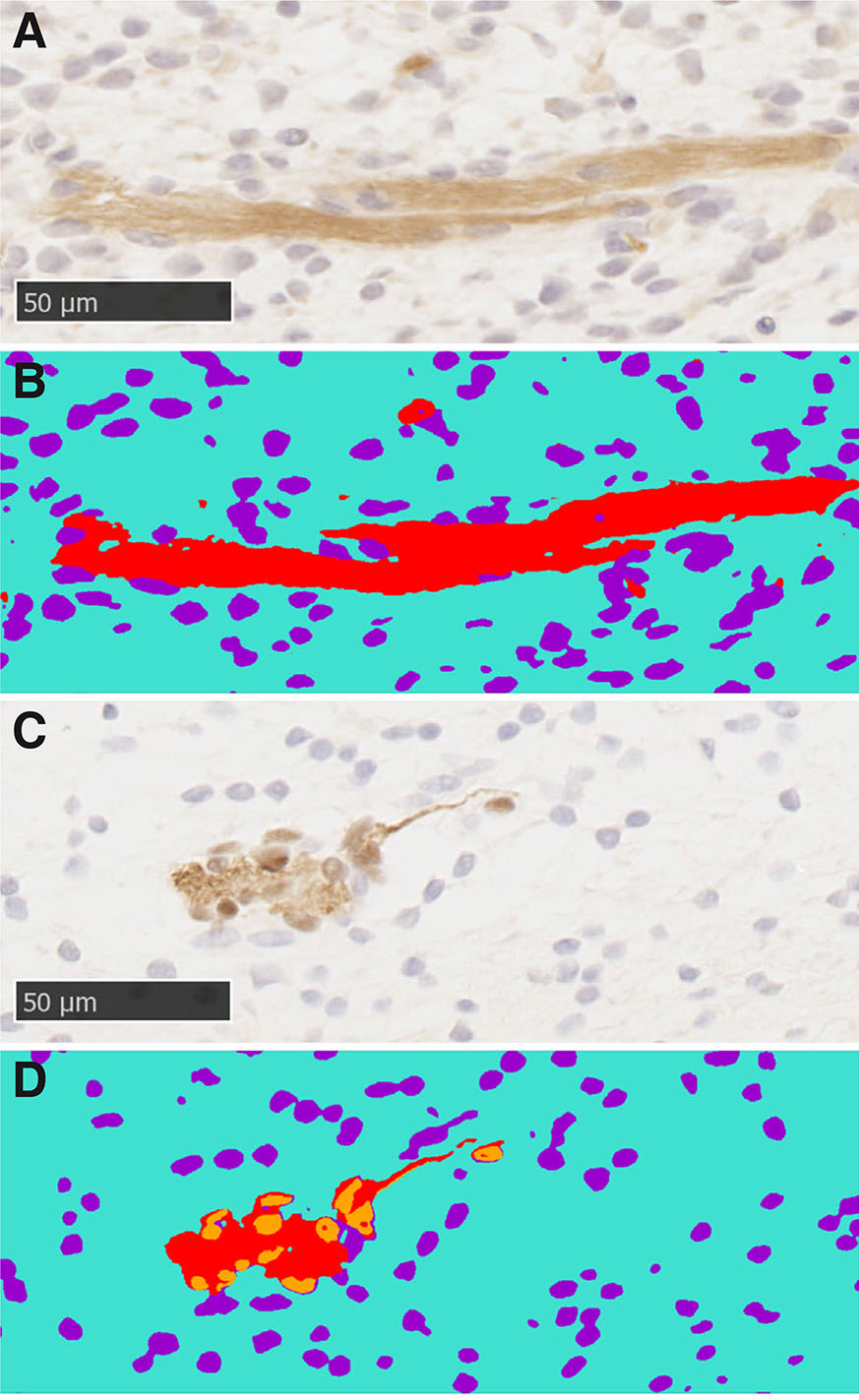
Fig. 13. Machine learning was used to segment images for analysis.
(A) Section stained with relaxin family peptide receptor 2 (RXFP2) antibody. (C) Section stained with SOX10 antibody. (B,D) Segmented images of A and C, respectively. Red, positively stained regions; orange, positively stained nuclei; purple, negative nuclei; cyan, background.
Statistical analysis
Statistical analyses were conducted in R (version 4.1.0). The paired Wilcoxon rank sum test was used to compare the horned measurements (InnerHB and OuterHB), and unpaired Wilcoxon rank sum tests were used to compare the horned and polled samples. To analyse the IHC data, the Fisher’s exact test was used to compare horned HB samples and Polled HB+FS samples.
Supplementary Material
Acknowledgements
JEA is funded by Meat & Livestock Australia. We would like to thank the team at the Roseworthy Production Animal Clinic for their veterinary care of the animals, in particular, Dr. Jamie Moffat for carrying out the laparoscopy surgeries. Tissue processing was carried out by the Histology Services at the Adelaide Health and Medical Sciences. Dr. Agatha Labrinidis and Dr. Jane Sibbons from Adelaide Microscopy are thanked for their guidance in using the histological equipment.
Abbreviations
HB, horn bud ; FS, frontal skin ; H&E, hematoxylin and eosin ; IHC, immunohistochemistry ;Declarations
Conflicts of interest
The authors declare no conflict of interest.
Data Availability
Code for data (10.25909/21753890 and 10.25909/24328915) and statistical analysis can be found on figshare (10.25909/19971098 and 10.25909/19972088).
References
Aldersey J. E., Sonstegard T. S., Williams J. L., Bottema C. D. K. (2020). Understanding the effects of the bovine POLLED variants. Animal Genetics 51: 166-176.
Allais-Bonnet A., Grohs C., Medugorac I., Krebs S., Djari A., Graf A., Fritz S., Seichter D., Baur A., Russ I., Bouet S., Rothammer S., Wahlberg P., Esquerré D., Hoze C., Boussaha M., Weiss B., Thépot D., Fouilloux M.N., Rossignol M.N., van Marle-Köster E., Hreiðarsdóttir G. E., Barbey S., Dozias D., Cobo E., Reversé P., Catros O., Marchand J.L., Soulas P., Roy P., Marquant-Leguienne B., Le Bourhis D., Clément L., Salas-Cortes L., Venot E., Pannetier M., Phocas F., Klopp C., Rocha D., Fouchet M., Journaux L., Bernard-Capel C., Ponsart C., Eggen A., Blum H., Gallard Y., Boichard D., Pailhoux E., Capitan A. (2013). Novel Insights into the Bovine Polled Phenotype and Horn Ontogenesis in Bovidae. PLoS ONE 8: e63512.
Buda S., Bragulla H., Budras K. D., (2011). Central nervous system and cranial nerves. In Bovine anatomy. (Ed. Budras K. D., ) Schlutersche, Hannover, Germany.
Capitan A., Allais-Bonnet A., Pinton A., Marquant-Le Guienne B., Le Bourhis D., Grohs C., Bouet S., Clément L., Salas-Cortes L., Venot E., Chaffaux S., Weiss B., Delpeuch A., Noé G., Rossignol M.N., Barbey S., Dozias D., Cobo E., Barasc H., Auguste A., Pannetier M., Deloche M.C., Lhuilier E., Bouchez O., Esquerré D., Salin G., Klopp C., Donnadieu C., Chantry-Darmon C., Hayes H., Gallard Y., Ponsart C., Boichard D., Pailhoux E. (2012). A 3.7 Mb Deletion Encompassing ZEB2 Causes a Novel Polled and Multisystemic Syndrome in the Progeny of a Somatic Mosaic Bull. PLoS ONE 7: e49084.
Carlson D. F., Lancto C. A., Zang B., Kim E.S., Walton M., Oldeschulte D., Seabury C., Sonstegard T. S., Fahrenkrug S. C. (2016). Production of hornless dairy cattle from genome-edited cell lines. Nature Biotechnology 34: 479-481.
Duijvesteijn N., Bolormaa S., Daetwyler H. D., van der Werf J. H. J. (2018). Genomic prediction of the polled and horned phenotypes in Merino sheep. Genetics Selection Evolution 50: 28.
Evans H. E., Sack W. O. (1973). Prenatal Development of Domestic and Laboratory Mammals: Growth Curves, External Features and Selected References. Anatomia, Histologia, Embryologia: Journal of Veterinary Medicine Series C 2: 11-45.
Fuchs E. (2007). Scratching the surface of skin development. Nature 445: 834-842.
Goncharuk S. A., Artemieva L. E., Nadezhdin K. D., Arseniev A. S., Mineev K. S. (2020). Revising the mechanism of p75NTR activation: intrinsically monomeric state of death domains invokes the "helper" hypothesis. Scientific Reports 10: 13686.
He S., Di J., Han B., Chen L., Liu M., Li W. (2020). Genome-Wide Scan for Runs of Homozygosity Identifies Candidate Genes Related to Economically Important Traits in Chinese Merino. Animals 10: 524.
Horikiri T., Ohi H., Shibata M., Ikeya M., Ueno M., Sotozono C., Kinoshita S., Sato T. (2017). SOX10-Nano-Lantern Reporter Human iPS Cells; A Versatile Tool for Neural Crest Research. PLOS ONE 12: e0170342.
Johnston S., McEwan J. C., Pickering N., Kijas J. W., Beraldi D., Pilkington J. G., Pemberton J. M., Slate J. (2011). Genome‐wide association mapping identifies the genetic basis of discrete and quantitative variation in sexual weaponry in a wild sheep population. Molecular Ecology 20: 2555-2566.
Kardos M., Luikart G., Bunch R., Dewey S., Edwards W., McWilliam S., Stephenson J., Allendorf F. W., Hogg J. T., Kijas J. (2015). Whole‐genome resequencing uncovers molecular signatures of natural and sexual selection in wild bighorn sheep. Molecular Ecology 24: 5616-5632.
Kim J., Lo L., Dormand E., Anderson D. J. (2003). SOX10 Maintains Multipotency and Inhibits Neuronal Differentiation of Neural Crest Stem Cells. Neuron 38: 17-31.
Kling-Eveillard F., Knierim U., Irrgang N., Gottardo F., Ricci R., Dockès A.C. (2015). Attitudes of farmers towards cattle dehorning. Livestock Science 179: 12-21.
Li J., Chatzeli L., Panousopoulou E., Tucker A. S., Green J. B. A. (2016). Epithelial stratification and placode invagination are separable functions in early morphogenesis of the molar tooth. Development 143: 670-681.
Lu B., Pang P. T., Woo N. H. (2005). The yin and yang of neurotrophin action. Nature Reviews Neuroscience 6: 603-614.
Lühken G., Krebs S., Rothammer S., Küpper J., Mioč B., Russ I., Medugorac I. (2016). The 1.78-kb insertion in the 3′-untranslated region of RXFP2 does not segregate with horn status in sheep breeds with variable horn status. Genetics Selection Evolution 48: 78.
Mintline E. M., Stewart M., Rogers A. R., Cox N. R., Verkerk G. A., Stookey J. M., Webster J. R., Tucker C. B. (2013). Play behavior as an indicator of animal welfare: Disbudding in dairy calves. Applied Animal Behaviour Science 144: 22-30.
Nesbit M., Mamo J. C., Majimbi M., Lam V., Takechi R. (2021). Automated Quantitative Analysis of ex vivo Blood-Brain Barrier Permeability Using Intellesis Machine-Learning. Frontiers in Neuroscience 15: 617221.
Pan Z., Li S., Liu Q., Wang Z., Zhou Z., Di R., Miao B., Hu W., Wang X., Hu X., Xu Z., Wei D., He X., Yuan L., Guo X., Liang B., Wang R., Li X., Cao X., Dong X., Xia Q., Shi H., Hao G., Yang J., Luosang C., Zhao Y., Jin M., Zhang Y., Lv S., Li F., Ding G., Chu M., Li Y. (2018). Whole-genome sequences of 89 Chinese sheep suggest role of RXFP2 in the development of unique horn phenotype as response to semi-feralization. GigaScience 7: giy019.
Puthiyaveetil J. S., Kota K., Chakkarayan R., Chakkarayan J., Thodiyil A. K., (2016). Epithelial - Mesenchymal Interactions in Tooth Development and the Significant Role of Growth Factors and Genes with Emphasis on Mesenchyme - A Review. Journal of clinical and diagnostic research : JCDR 10: ZE05-ZE09.
Rapizzi E., Benvenuti S., Deledda C., Martinelli S., Sarchielli E., Fibbi B., Luciani P., Mazzanti B., Pantaleo M., Marroncini G., Vannelli G. B., Maggi M., Mannelli M., Luconi M., Peri A. (2020). A unique neuroendocrine cell model derived from the human foetal neural crest. Journal of Endocrinological Investigation 43: 1259-1269.
Salhotra A., Shah H. N., Levi B., Longaker M. T. (2020). Mechanisms of bone development and repair. Nature Reviews Molecular Cell Biology 21: 696-711.
Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9: 676-682.
Schuster F., Aldag P., Frenzel A., Hadeler K.G., Lucas-Hahn A., Niemann H., Petersen B. (2020). CRISPR/Cas12a mediated knock-in of the Polled Celtic variant to produce a polled genotype in dairy cattle. Scientific Reports 10: 13570.
Simon R., Drögemüller C., Lühken G. (2022). The Complex and Diverse Genetic Architecture of the Absence of Horns (Polledness) in Domestic Ruminants, including Goats and Sheep. Genes 13: 832.
Svandova E., Peterkova R., Matalova E., Lesot H. (2020). Formation and Developmental Specification of the Odontogenic and Osteogenic Mesenchymes. Frontiers in Cell and Developmental Biology 8: 640.
Sylvester S. P., Stafford K. J., Mellor D.J., Bruce R. A., Ward R. N. (2004). Behavioural responses of calves to amputation dehorning with and without local anaesthesia. Australian Veterinary Journal 82: 697-700.
- Wang G., Gill C.A., (2021). Dissection of the scurs phenotype to refine the mapping of scurs. Texas Agricultural and Mechanical University, Poster presented at: International Society for Animal Genetics conference, July 26-30, 2021, [Online]
Wang H., Zhu H., Hu Z., Heng N., Gong J., Wang Y., Zou H., Zhao S. (2022). Uncovering Novel Features of the Pc Locus in Horn Development from Gene-Edited Holstein Cattle by RNA-Sequencing Analysis. International Journal of Molecular Sciences 23: 12060.
Wang Y., Zhang C., Wang N., Li Z., Heller R., Liu R., Zhao Y., Han J., Pan X., Zheng Z., Dai X., Chen C., Dou M., Peng S., Chen X., Liu J., Li M., Wang K., Liu C., Lin Z., Chen L., Hao F., Zhu W., Song C., Zhao C., Zheng C., Wang J., Hu S., Li C., Yang H., Jiang L., Li G., Liu M., Sonstegard T. S., Zhang G., Jiang Y., Wang W., Qiu Q. (2019). Genetic basis of ruminant headgear and rapid antler regeneration. Science 364: eaav6335.
Wiedemar N., Drögemüller C. (2015). A 1.8‐kb insertion in the 3′‐UTR of RXFP2 is associated with polledness in sheep . Animal Genetics 46: 457-461.
Wiedemar N., Tetens J., Jagannathan V., Menoud A., Neuenschwander S., Bruggmann R., Thaller G., Drögemüller C. (2014). Independent Polled Mutations Leading to Complex Gene Expression Differences in Cattle. PLoS ONE 9: e93435.
Wiener D. J., Wiedemar N., Welle M. M., Drögemüller C. (2015). Novel Features of the Prenatal Horn Bud Development in Cattle (Bos taurus). PLOS ONE 10: e0127691.