Int. J. Dev. Biol. 67: 19 - 25 (2023)
Single-cell transcriptome profiling reveals distinct expression patterns among genes in the mouse incisor dental pulp
Developmental Expression Pattern | Published: 29 March 2023
Abstract
SOX transcription factors play key roles in cell differentiation and cell fate determination during development. Using single-cell RNA-sequencing data, we examined the expression profiles of Sox genes in the mouse incisor dental pulp. Our analysis showed that Sox4, Sox5, Sox9, Sox11, and Sox12 are mainly expressed in mesenchymal stem/stromal cells (MSCs) representing osteogenic cells at different stages of differentiation. We found that in several MSCs, Sox genes co-expressed with regulatory genes such as Sp7, Satb2, Msx1, Snai2, Dlx1, Twist2, and Tfap2a. In addition, Sox family genes colocalized with Runx2 and Lef1, which are highly enriched in MSCs undergoing osteoblast differentiation. A protein interaction network analysis uncovered that CREBBP, CEBPB, TLE1, TWIST1, and members of the HDAC and SMAD families are interacting partners of RUNX2 and LEF1 during skeletal development. Collectively, the distinct expression patterns of the SOX transcription factors suggest that they play essential regulatory roles in directing lineage-specific gene expression during differentiation of MSCs.
Keywords
dental pulp, single cells, chromatin, incisors, transcriptome
Introduction
SOX transcription factors (TFs) are key regulators of stem cell maintenance, differentiation and cell fate specification (Liu and Guo, 2021; Stepanovic et al., 2021; Lefebvre, 2019). Several members of the SOX family play an essential role in the development of the neural crest, which is important for the formation of the craniofacial region (Schock and LaBonne, 2020). Because SOX TFs contribute to the determination of cell fate and lineage specification, mutations within this family often have been associated with birth defects. For instance, mutations in SOX9 lead to campomelic dysplasia, a severe disorder that affects development of the skeleton, reproductive system, and other parts of the body and heterozygous mutations of SOX10 cause multiple defects associated with malfunction of neural crest derivatives including melanocytes, the enteric nervous system, Schwann cells, oligodendrocytes and olfactory cells (Angelozzi and Lefebvre, 2019; Ming et al., 2022; Pingault et al., 2022).
Interestingly, recent research showed that SOX9 behaves like a pioneer TF by establishing accessible chromatin regions (Fuglerud et al., 2022). Genome-wide studies have revealed that sequentially expressed SOX TFs bind to the specific regulatory elements to coordinate lineage commitment (Klum et al., 2018). Single-cell transcriptomic studies identified Sox genes as key regulators of neuroglandular lineages in the cnidarians (Steger et al., 2022). In human pluripotent stem cells, SOX TFs establish a permissive chromatin landscape to activate the β-catenin-dependent Wingless-Int (WNT) signaling pathway (Mukherjee et al., 2022).
Although the function and regulatory mode of SOX TFs is well established in skeletogenesis and osteoblast and chondrocyte differentiation (Lefebvre, 2019), very little is known about their role in odontogenesis and dentin development. Previous work revealed that Sox genes exhibit a dynamic spatial and temporal expression pattern during mouse tooth development (Kawasaki et al., 2015). In the current study, we used single-cell RNA-sequencing (RNA-seq) data to demonstrate specific and distinct expression profiles of Sox genes in the mouse incisor dental pulp.
Results
Single-cell RNA expression of Sox genes in different cell subpopulations of the mouse incisor dental pulp
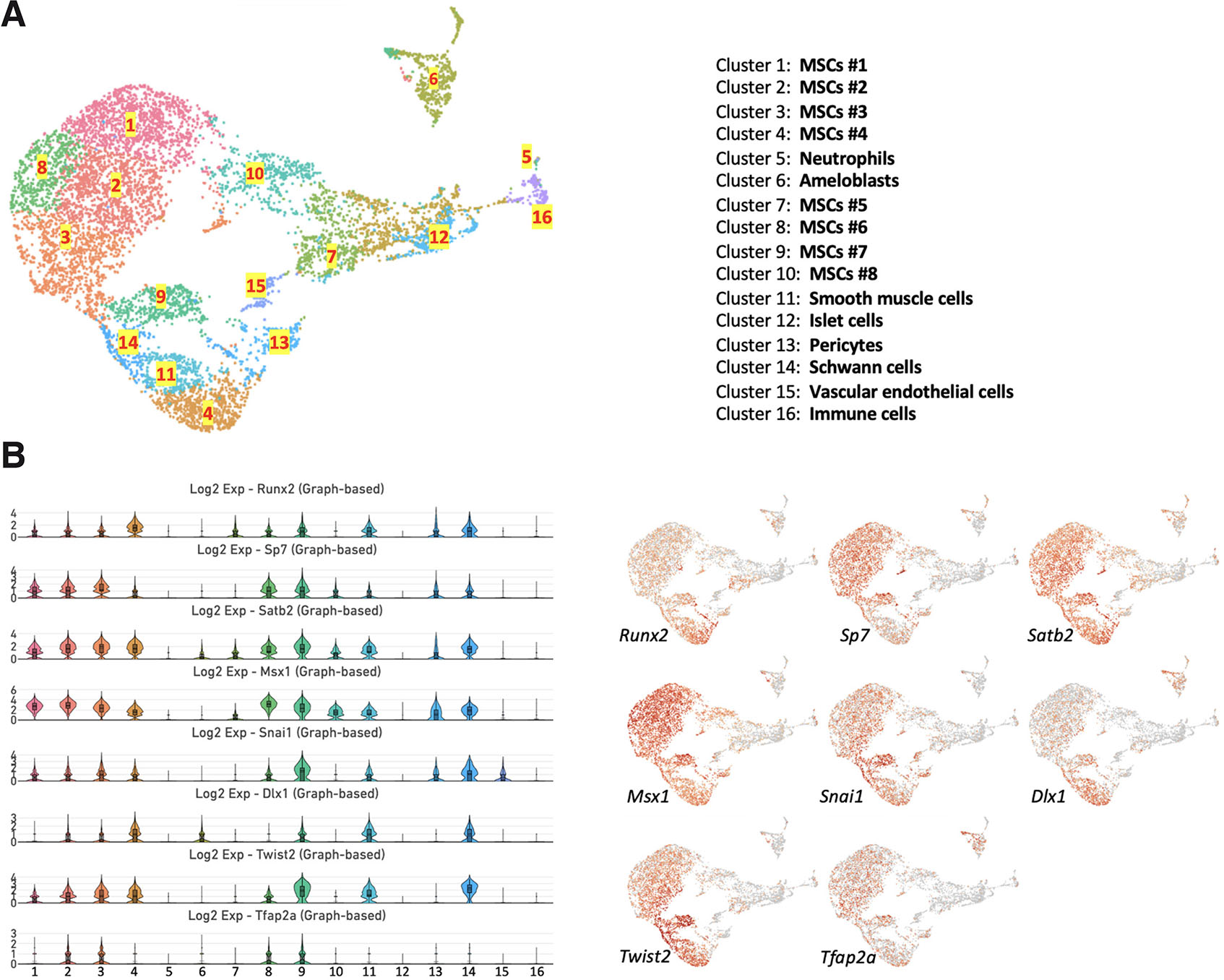
We previously identified 16 distinct cell clusters in the dental pulp of mouse incisors based on the differential expression of marker genes (Bayarsaihan et al., 2022). According to the expression of key genes required for osteogenic differentiation, we identified eight clusters of MSCs (Fig. 1A). Differential expression patterns of Runx2, Sp7, Satb2, Msx1, Snai1, Dlx1, Twist2, and Tfap2a suggest that these MSCs represent osteogenic subpopulations at various stages of differentiation (Fig. 1B).
Fig. 1. Expression of genes encoding osteogenic transcription factors.
(A) Uniform manifold approximation and projection (UMAP) visualization depicts the single-cell RNA expression profile of 16 cell types in the incisor dental pulp. (B) Violin plots (left) and UMAP visualization (right) of the Runx2, Sp7, Satb2, Msx1, Snai1, Dlx1, Twist2, and Tfap2a genes.
Table 1
The expression pattern of Sox genes in the mouse incisor dental pulp
| MSCs #1 | MSCs #2 | MSCs #3 | MSCs #4 | Ameloblasts | MSCs #5 | MSCs #6 | MSCs #7 | MSCs #8 | Smooth muscle cells | |
|---|---|---|---|---|---|---|---|---|---|---|
| Sox4 | + | + | + | + | + | + | + | + | + | + |
| Sox5 | + | + | + | + | + | |||||
| Sox9 | + | + | + | + | + | + | + | + | + | + |
| Sox11 | + | + | + | + | + | + | + | + | + | + |
| Sox12 | + | + | + | + | + | + |
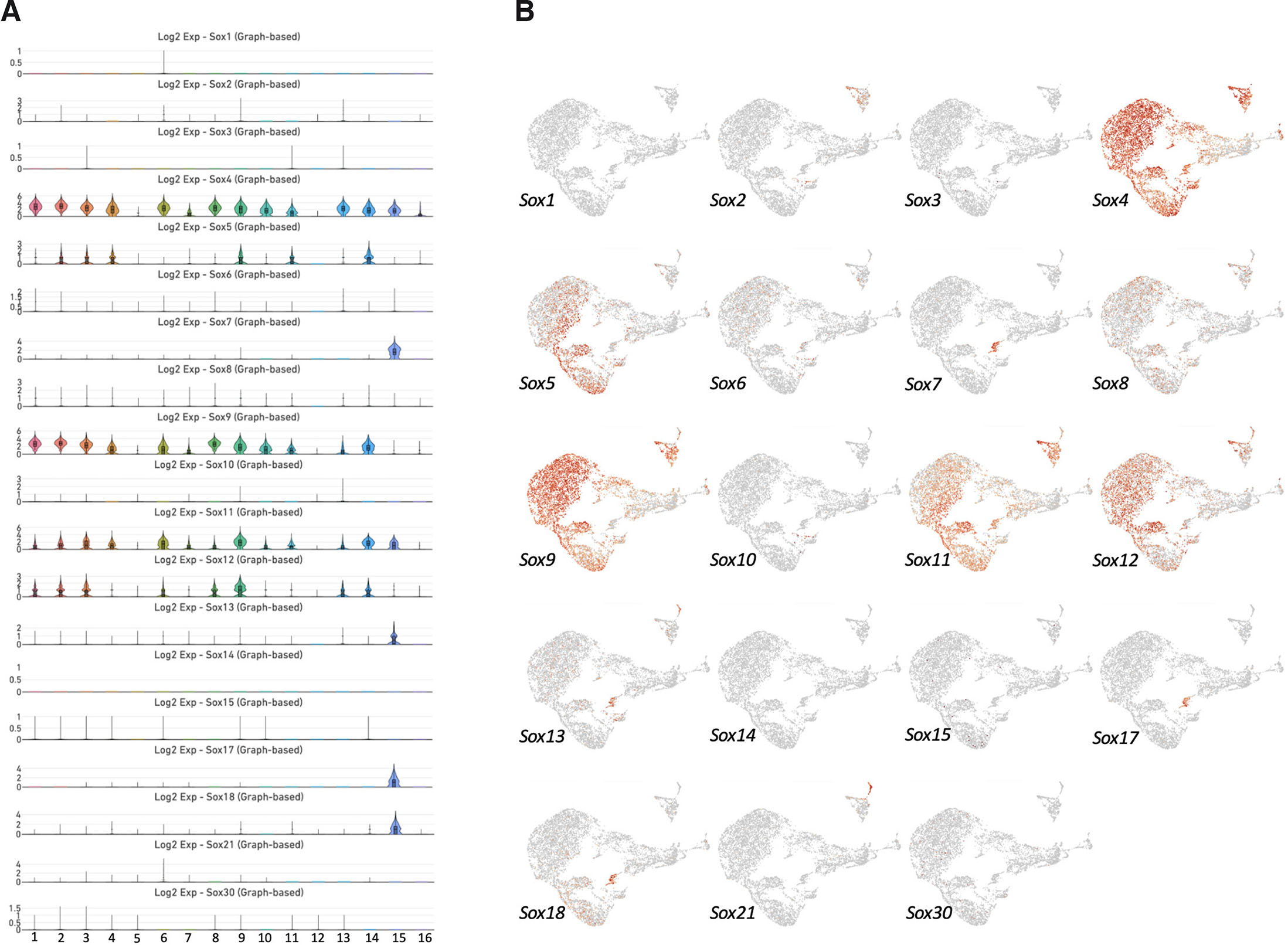
We found that Sox1, Sox2, Sox3, Sox6, Sox8, Sox10, Sox15, Sox21, and Sox30 exhibited weak expression in the mouse incisor dental pulp (Fig. 2). Interestingly, our single-cell RNA-seq failed to detect expression of Sox14 in any of the dental pulp subpopulations. By contrast, Sox4 was vigorously expressed in clusters 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 13, 14, 15, and 16; Sox5 in clusters 2, 3, 4, 9, 11, and 14; Sox9 in clusters 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 13, and 14; Sox11 in clusters 1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 13, 14, and 15; and Sox12 in clusters 1, 2, 3, 6, 8, 9, 13, and 14. Interestingly, Sox7, Sox13, Sox17, and Sox18 were mainly enriched in only a single cluster, cluster 15.
Fig. 2. Expression of genes encoding members of the SOX family of transcription factors.
(A) Violin plots and (B) UMAP visualization revealed that Sox4, Sox5, Sox9, Sox11, and Sox12 are mainly expressed in mesenchymal stem/stromal cells (MSCs), whereas Sox7, Sox13, Sox17, and Sox18 are enriched in vascular endothelial cells.
Chromatin accessibility and 5hmC of Sox genes in the mouse incisor dental pulp
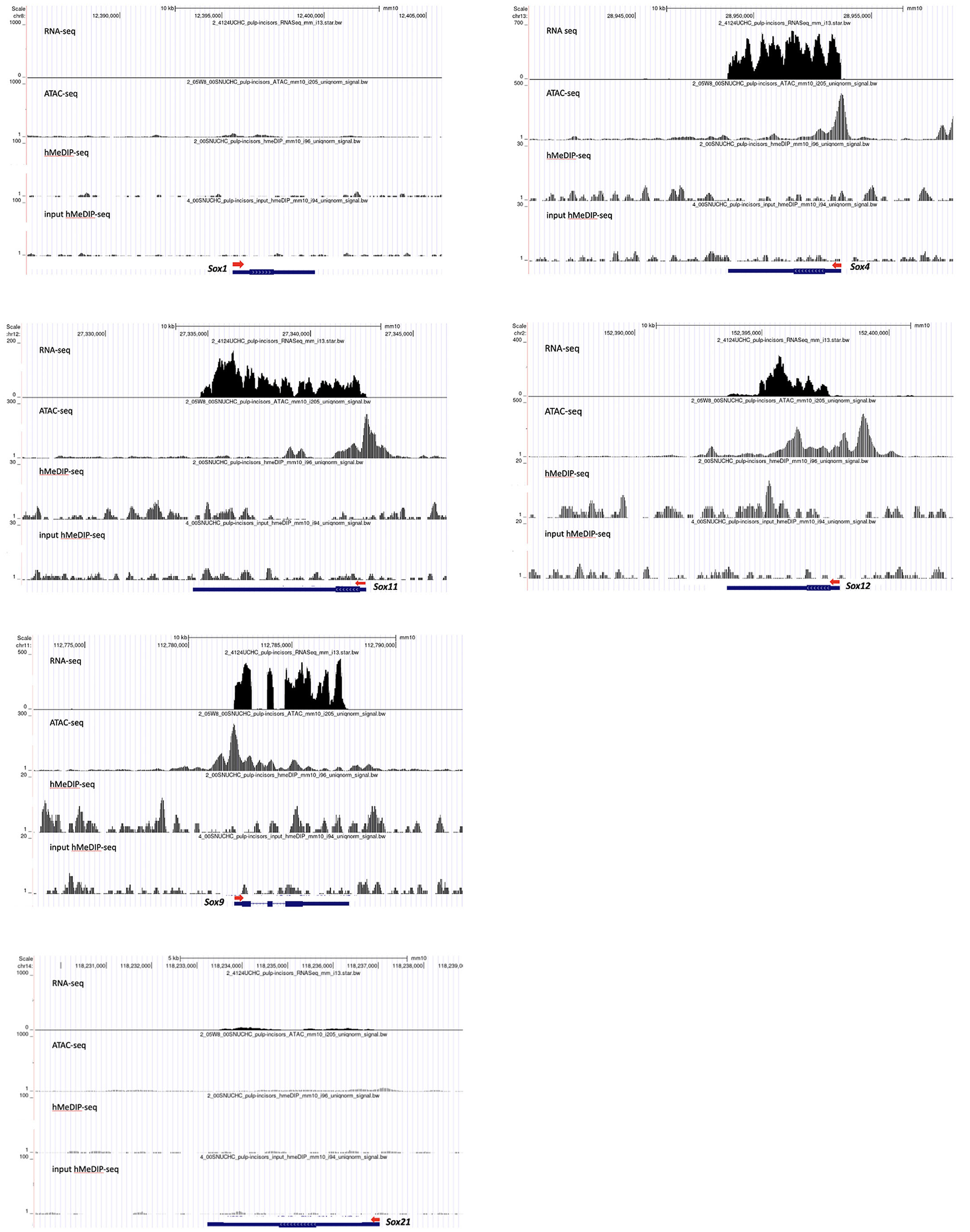
Genomic regions of actively transcribed genes retain open chromatin and are characterized by the enrichment of 5hmC, a hallmark feature of DNA demethylation. Hence, we investigated chromatin accessibility and DNA demethylation by performing transposase-accessible chromatin sequencing (ATAC-seq) and hydroxymethylation DNA immunoprecipitation sequencing (hMeDIP-seq). Using these assays, we demonstrated that actively transcribed Sox genes retain characteristics of open chromatin. Transcriptome profiling using bulk RNA-seq showed that Sox4, Sox9, Sox11, and Sox12 are strongly expressed in the molar dental pulp (Fig. 3). We observed elevated ATAC-seq peaks within the transcription start sites and promoter regions of these genes as well as within the gene body regions. Using hMeDIP-seq, we detected 5hmC enrichment across the gene bodies and the proximal and distal regions of Sox4, Sox9, Sox11, and Sox12, whereas input hMeDIP-seq showed low levels of 5hmC. We also analyzed the genomic regions of Sox1 and Sox21, which displayed very weak or no expression in dental pulp. Both genes retained closed chromatin and we failed to detect 5hmC marks along the genomic regions of these genes. Collectively, our results support the previous observation that genes associated with cell differentiation and lineage commitment retain an open chromatin configuration and are enriched in 5hmC in the mouse dental pulp (Pujan et al., 2022a; Pujan et al., 2002b).
Fig. 3. Fig. 3. Chromatin accessibility, 5-hydroxymethylcytosine (5hmC) landscape and bulk RNA expression of Sox genes.
Sox1 and Sox21 were not expressed in the mouse incisor dental pulp. We failed to detect ATAC-seq peaks or enrichment of 5hmC in the genomic regions of Sox1 and Sox21. In contrast, strong ATAC-seq peaks were present at the transcription start site, promoter regions, and gene bodies of Sox4, Sox9, Sox11, and Sox12. The 5hmC peaks were also high across the genomic regions of these genes.
Single-cell RNA expression profile of interacting partners of Sox genes in MSCs of the mouse incisor dental pulp
Our research demonstrated that only a few members of the Sox gene family, specifically Sox4, Sox9, Sox11, and Sox12, retain a highly specific expression pattern within different clusters of the mouse incisor dental pulp (Fig. 2). SOX4, SOX11, and SOX12 comprise the SOXC group of TFs, which play key functions in cell fate determination (Lefebvre and Bhattaram, 2016). Conversely, SOX9 is essential for chondrocyte fate maintenance and differentiation (Lefebvre, 2019).
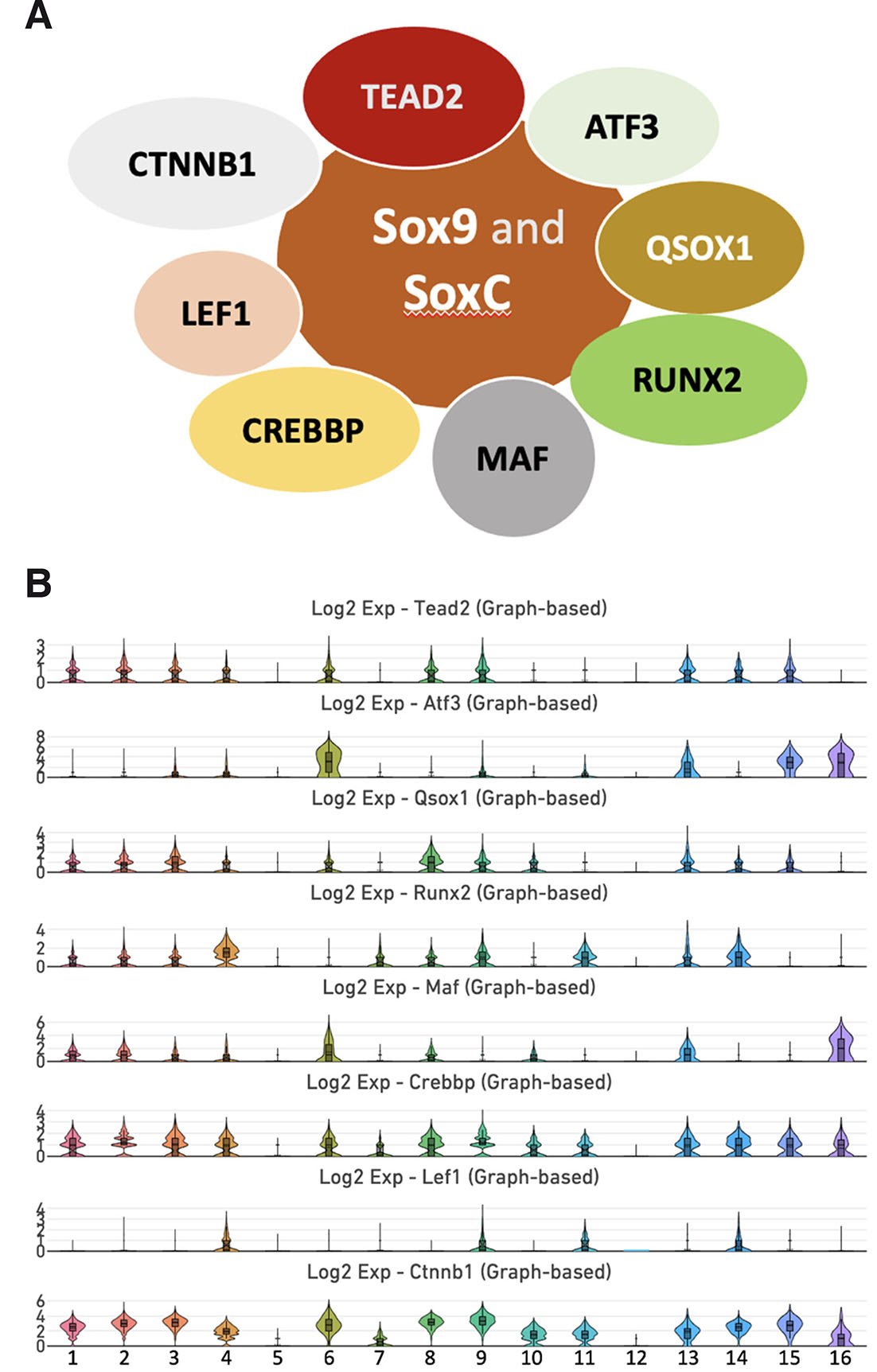
By analyzing the STRING interaction network (https://string-db.org), we discovered that the SOXC group and SOX9 have a specific set of interacting partners (Fig. 4). Among these interacting partners, we identified TEAD2, ATF3, QSOX1, RUNX2, MAF, CREBBP, LEF1, and CTNNB1. We found that within MSC subpopulations, Tead2 was enriched in clusters 1, 2, 3, 4, 8, and 9, while Atf3 was weakly expressed in clusters 3, 4, and 9. Qsox1 encodes an enzyme that catalyzes disulfide bond formation and is involved in redox regulation (Reznik and Fass, 2022). In our analysis, Qsox1 was expressed in MSC clusters 1, 2, 3, 4, 8, 9, and 10; Runx2 in cluster 1, 2, 3, 4, 7, 8, and 9; and Maf in clusters 1, 2, 3, 4, 8, and 10. Lef1 exhibited a very restricted expression pattern in clusters 4 and 9. Both Crebbp and Ctnnb1 were vigorously expressed in clusters 1, 2, 3, 4, 7, 8, 9, and 10.
Fig. 4. Interaction network of SOX9 and SOXC and single-cell transcriptome clustering analysis.
(A) STRING interaction network analysis revealed that SOX factors associate with TEAD2, ATF3, QSOX1, RUNX2, MAF, CREBBP, LEF1, and CTNNB1. (B) Violin plots revealed that Tead2, Qsox1, Runx2, Crebbp, and Ctnnb1 co-express with Sox9 and SoxC group in the MSC clusters 1, 2, 3, 4, 8, and 9. The expression of Atf3 was limited to clusters 3, 4, and 9, whereas Maf exhibited wide expression in clusters 1, 2, 3, 4, 7, 8, and 9. Lef1 was enriched in clusters 4 and 9.
Single-cell RNA expression profile of interacting partners of Runx2 and Lef1 in MSCs of the mouse incisor dental pulp
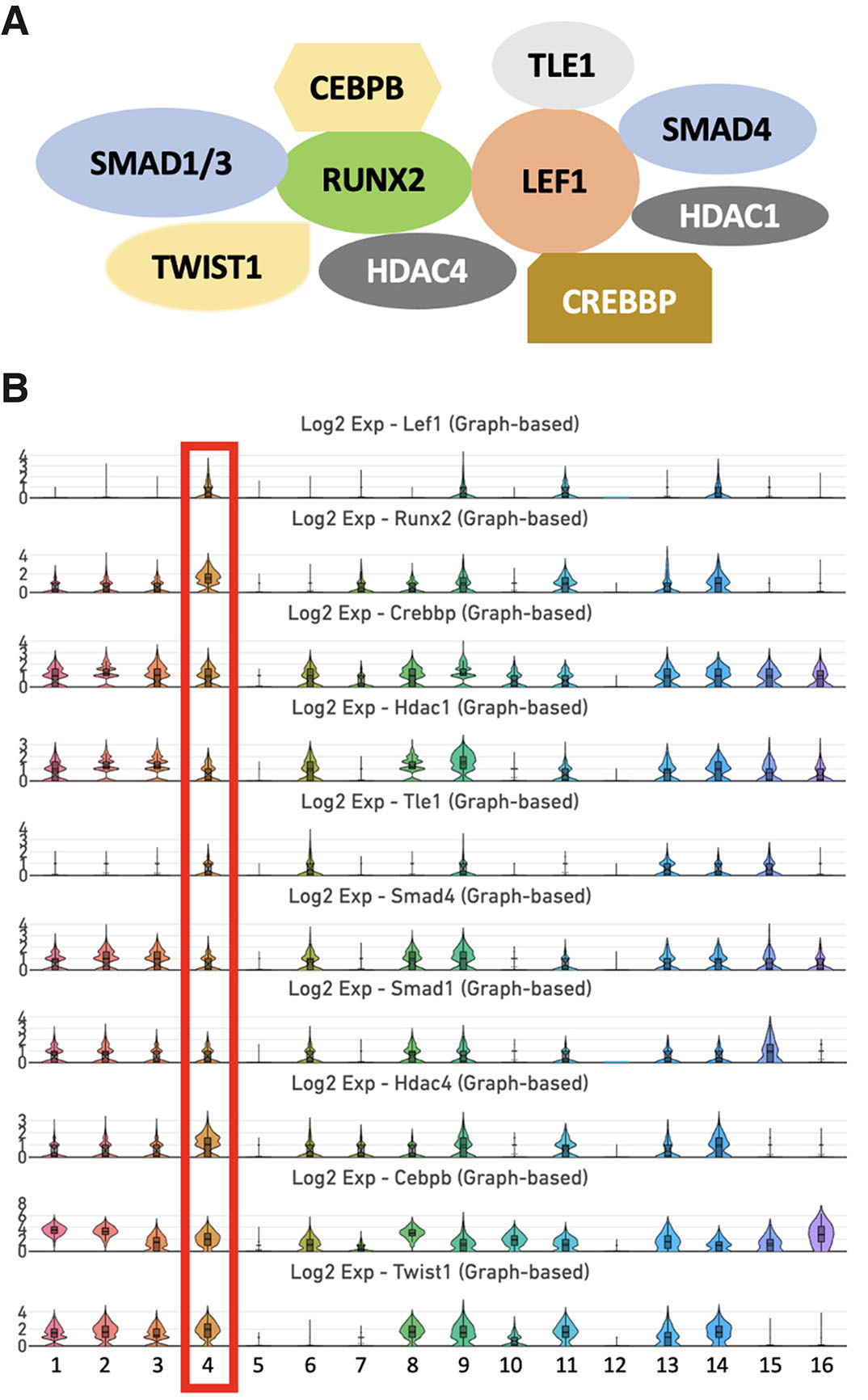
The expression of Runx2 and Lef1 was highly enriched in cluster 4, which represents the osteoblastic lineage. RUNX2 interacts with LEF1 to induce canonical WNT signaling during skeletal development (Reinhold and Naski, 2007). Therefore, we analyzed the expression pattern of interacting partners of RUNX2 and LEF1 in cluster 4. Using the STRING interaction network, we uncovered the interacting partners of RUNX2 and LEF1 in the dental pulp (Fig. 5). We found that genes encoding several canonical interacting partners of RUNX2 and LEF1, such as Crebbp, Hdac1, Hdac4, Tle1, Smad1, Smad4, Cebpb, and Twist1, were enriched in cluster 4.
Fig. 5. Interaction network of RUNX2 and LEF1 and single-cell transcriptome clustering analysis.
(A) STRING interaction network analysis revealed that RUNX2 and LEF1 associate with TLE1, SMAD4, CREBBP, TWIST1, CEBPB, and members of the HDAC and SMAD families. (B) Violin plots showed that Crebbp, Hdac1, Hdac4, Tle1, Smad1, Smad4, Cebpb, and Twist1 co-express with Runx2 and Lef1 in cluster 4, which represents MSCs of the osteoblastic lineage.
Discussion
We previously identified several clusters of MSCs in the mouse incisor dental pulp that represent subpopulations of osteogenic cells at different stages of differentiation (Bayarsaihan et al., 2022). These MSCs express a wide range of genes including Msx1, Snai1, Dlx1, Twist2, and Tfap2a, that are important for differentiation of the neural crest, osteogenic, and chondrogenic lineages (Fig. 1B). Our current analysis showed that these genes are co-expressed with Sox4, Sox5, Sox9, Sox11, and Sox12 in the mouse incisor dental pulp.
SOX TFs represent key nodes in the gene regulatory networks governing stem cell maintenance, cellular differentiation and lineage commitment (Abdelalim et al., 2014; Lefebvre and Bhattaram, 2016; Lefebvre, 2019). For instance, high expression of SOXC TFs in osteodermal progenitors causes skull and skin underdevelopment (Angelozzi et al., 2022). Recent single-cell genomic studies revealed that SOX TFs orchestrate cell fate decisions during sensorineural regeneration in zebrafish (Jimenez et al., 2022). Combinatorial activity between SOX and SIX TFs underlies this regulatory network. During neuronal and glial differentiation, sequential activity of SOX TFs mediates the specification of neural lineages (Stevanovic et al., 2021). A recent study suggested that SOX11 is a novel regulator of odontoblast differentiation (Inoue et al., 2022). The regulatory mode of SOX TFs is achieved by forming functional interactions with other TFs and chromatin remodeling and epigenetic complexes (Weider and Wegner, 2017). In this regard, RUNX2 and LEF1 are essential components of gene regulatory cross-wirings during bone formation and homeostasis (Rodríguez-Carballo et al., 2011). It is well established that the interaction of RUNX2 with LEF1 triggers the canonical WNT signaling pathway during skeletal development (Reinhold and Naski, 2007). In our analysis, Runx2 and Lef1 were highly enriched in cluster 4, which represents MSCs undergoing osteoblastic differentiation. Our STRING interaction network analysis uncovered that both TFs interact with CREBBP, CEBPB, TLE1, TWIST1, and members of the histone deacetylases (HDAC) and SMAD families. Genes encoding these regulatory proteins were co-expressed with Runx2 and Lef1 in cluster 4. Crebbp encodes the CBP co-activator of the CBP/p300 histone acetyltransferase family, which plays an important role in MSC osteogenic and chondrogenic differentiation (Ma et al., 2022).
CCAAT/enhancer binding protein beta (C/EBPβ), encoded by Cebpb, is a critical regulator of both adipocyte and osteoblast differentiation (Motyl et al., 2011). The transcriptional corepressor transducin-like enhancer-1 (TLE1), a product of the Tle1 gene, functions with RUNX2 in the epigenetic suppression of ribosomal RNA genes (Ali et al., 2010). An interaction between TWIST1 and RUNX2 coordinates craniofacial muscle development (Han et al., 2021). Studies have shown that both the canonical and non-canonical SMAD-dependent signaling pathways control the proliferation and differentiation of dental MSCs as well as osteogenesis and skeletal development (Liu et al., 2022; Wu et al., 2016). Histone deacetylation via HDACs is an important function of the multiprotein epigenetic complexes that interact with many TFs, including RUNX2 and LEF1, during skeletal development (Jensen et al., 2010; Huynh et al., 2017).
Coordinated crosstalk among the bone morphogenic protein (BMP), fibroblast growth factor (FGF) and WNT signaling pathways from the dental epithelium to the mesenchyme underlies the regulation of the odontogenic regulatory network during early tooth development (Chen et al., 2009). BMP signaling utilizes the canonical and non-canonical SMAD signaling pathways, which engage different TFs including RUNX2 and SP7 to initiate the differentiation of MSCs along the odontoblastic lineage (Liu et al., 2022).
RUNX2 is an essential TF in osteogenic differentiation and early tooth development (Wen et al., 2020). In committed odontoblasts, RUNX2 is required for bone matrix protein gene expression (Qin et al., 2021). Depletion of the Runx2 gene in odontoblasts leads to shorter and irregularly aligned incisors. Wen et al., showed that Runx2 plays a crucial role in tooth root development and the differentiation of root progenitor cells. Ablation of Runx2 results in upregulation of canonical WNT signaling and interference of odontoblast differentiation (Wen et al., 2020). At the late stages of odontoblast differentiation and dentin formation, RUNX2 attenuates terminal differentiation of odontoblasts while enhancing differentiation of odontoblasts to osteoblasts (Li et al., 2011). Interestingly, SOX9 directly interacts with RUNX2 and inhibits its activity during skeletogenesis (Zhou et al., 2006).
It is well accepted that TFs exert their function by interacting with epigenetic enzymes, coactivators and chromatin remodeling complexes (Dubois-Chevalier et al., 2015; Hernandez-Hernandez et al., 2020). In eukaryotes, TFs form large regulatory domains known as super-enhancers that define cell identity by expressing cell-type-specific genes (Whyte et al., 2013). Although we know very little about the contribution of the SOX family to dentin formation, the modular nature of TFs in forming gene regulatory networks suggests that crosstalk between SOX factors and bone-associated TFs could underlie the odontoblast-specific regulatory program. For example, SOX5, SOX6, and SOX9 cooperate genome-wide through super-enhancers to regulate chondrogenesis (Liu and Lefebvre, 2015). During skeletal development, SOX9 cooperates with RUNX2 and SP7 to coordinate cell-fate specification of osteoblasts and chondrocytes (Ohba, 2021).
In summary, we determined distinct cell-specific expression patterns of Sox genes in the mouse incisor dental pulp. A few members of the Sox family colocalize with key regulatory genes encoding TFs, epigenetic enzymes, and signaling molecules that play significant roles during skeletal and dental development.
Materials and Methods
Single-cell RNA-seq, genome-wide ATAC-seq, hMeDIP-seq, and bulk RNA-seq
Primary pulp isolation and viability of each single-cell suspension was performed as previously described (Bayarsaihan et al., 2022; Enkhmandakh et al., 2022). Suspensions of dissociated cells were loaded for capture using a Chromium Single Cell 3′ Reagent kit, v2 Chemistry (10× Genomics). Following capture and lysis, complementary DNA was synthesized and amplified (14 cycles) as per the 10×Genomics protocol. The amplified cDNA was used to construct an Illumina sequencing library and was sequenced on a single lane of a HiSeq 4000 (Illumina, San Diego, CA). ATAC-seq, hMeDIP-seq, and bulk RNA-seq services were performed as previously described (Joshi et al., 2022a; Joshi et al., 2022b).
Computational analysis
For single-cell RNA-seq, we excluded reads with more than one mismatch in the 8-bp i7 index. Using the STAR aligner, we retained only reads with MAPQ scores greater than 255. We excluded reads containing bases with Q30 scores below 3. After alignment, cell barcodes were filtered (up to one mismatch) against a whitelist of 737,500 barcodes provided by 10×Genomics. Cell-associated barcodes were distinguished from those associated with ambient mRNA using an adaptively computed unique molecular identifier threshold. After this filtering step, a digital counts matrix for the pulp sample was generated. The uniform manifold approximation and projection (UMAP), batch correction software BBKNN and FASTQ generation, MACS2 peak-calling algorithm, STAR aligner, and feature counts (FPKM assignment to genes) were performed as previously described (Bayarsaihan et al., 2022; Enkhmandakh et al., 2022).
Acknowledgements
This project was funded by the grant REP-401754-20144-20 from the University of Connecticut to DB. The data that support the findings of this study are available on request.
Abbreviations
5hmC, 5-hydroxymethylcytosine ;Declarations
Author contributions
DB: conceptualization, funding acquisition and supervision. BE: methodology. DB and BE: data analysis and writing. All authors gave final approval and agreed to be accountable for all aspects of the work.
References
Abdelalim E. M., Emara M. M., Kolatkar P. R. (2014). The SOX Transcription Factors as Key Players in Pluripotent Stem Cells. Stem Cells and Development 23: 2687-2699.
Ali S. A., Zaidi S. K., Dobson J. R., Shakoori A. R., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2010). Transcriptional corepressor TLE1 functions with Runx2 in epigenetic repression of ribosomal RNA genes. Proceedings of the National Academy of Sciences 107: 4165-4169.
Angelozzi M., Lefebvre V. (2019). SOXopathies: Growing Family of Developmental Disorders Due to SOX Mutations. Trends in Genetics 35: 658-671.
Angelozzi M., Pellegrino da Silva R., Gonzalez M. V., Lefebvre V. (2022). Single-cell atlas of craniogenesis uncovers SOXC-dependent, highly proliferative, and myofibroblast-like osteodermal progenitors. Cell Reports 40: 111045.
Bayarsaihan D., Enkhmandakh B., Vijaykumar A., Robson P., Mina M. (2022). Single-cell transcriptome analysis defines mesenchymal stromal cells in the mouse incisor dental pulp. Gene Expression Patterns 43: 119228.
Dubois-Chevalier J., Staels B., Lefebvre P., Eeckhoute J. (2015). The ubiquitous transcription factor CTCF promotes lineage-specific epigenomic remodeling and establishment of transcriptional networks driving cell differentiation. Nucleus 6: 15-18.
Enkhmandakh B., Robson P., Joshi P., Vijaykumar A., Shin D.G., Mina M., Bayarsaihan D. (2022). Single-Cell Transcriptome Analysis Defines Expression of Kabuki Syndrome-Associated KMT2D Targets and Interacting Partners. Stem Cells International 2022: 1-9.
Fuglerud B. M., Drissler S., Lotto J., Stephan T. L., Thakur A., Cullum R., Hoodless P. A. (2022). SOX9 reprograms endothelial cells by altering the chromatin landscape. Nucleic Acids Research 50: 8547-8565.
Chen J., Lan Y., Baek J.A., Gao Y., Jiang R. (2009). Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Developmental Biology 334: 174-185.
Han X., Feng J., Guo T., Loh Y.H. E., Yuan Y., Ho T.V., Cho C. K., Li J., Jing J., Janeckova E., He J., Pei F., Bi J., Song B., Chai Y. (2022). Runx2-Twist1 interaction coordinates cranial neural crest guidance of soft palate myogenesis. eLife 10: e62387.
Hernández-Hernández O., Ávila-Avilés R. D., Hernández-Hernández J. M. (2020). Chromatin Landscape During Skeletal Muscle Differentiation. Frontiers in Genetics 11: 578712.
Huynh N. C.N., Everts V., Ampornaramveth R. S. (2017). Histone deacetylases and their roles in mineralized tissue regeneration. Bone Reports 7: 33-40.
Inoue K., Matsuzaka K., Inoue T. (2022). Identification of Novel Regulator Involved in Differentiation of Mouse iPS Cells into Odontoblast-like Cells. The Bulletin of Tokyo Dental College 63: 119-128.
Jensen E. D., Gopalakrishnan R., Westendorf J. J. (2010). Regulation of gene expression in osteoblasts. BioFactors 36: 25-32.
Jimenez E., Slevin C. C., Song W., Chen Z., Frederickson S. C., Gildea D., Wu W., Elkahloun A. G., Ovcharenko I., Burgess S. M. (2022). A regulatory network of Sox and Six transcription factors initiate a cell fate transformation during hearing regeneration in adult zebrafish. Cell Genomics 2: 100170.
Joshi P., Vijaykumar A., Enkhmandakh B., Mina M., Shin D.G., Bayarsaihan D. (2022a). Genome-wide distribution of 5hmC in the dental pulp of mouse molars and incisors. The Journal of Biochemistry 171: 123-129.
Joshi P., Vijaykumar A., Enkhmandakh B., Shin D.G., Mina M., Bayarsaihan D. (2022b). The chromatin accessibility landscape in the dental pulp of mouse molars and incisors. Acta Biochimica Polonica 69: 131-138.
Lefebvre V., Bhattaram P. (2016). SOXC Genes and the Control of Skeletogenesis. Current Osteoporosis Reports 14: 32-38.
Lefebvre V. (2019). Roles and regulation of SOX transcription factors in skeletogenesis. In Vertebrate Skeletal Development. Elsevier.
Li S., Kong H., Yao N., Yu Q., Wang P., Lin Y., Wang J., Kuang R., Zhao X., Xu J., Zhu Q., Ni L. (2011). The role of runt-related transcription factor 2 (Runx2) in the late stage of odontoblast differentiation and dentin formation. Biochemical and Biophysical Research Communications 410: 698-704.
Liu C.F., Lefebvre V. (2015). The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Research 43: 8183-8203.
Liu M., Goldman G., MacDougall M., Chen S. (2022). BMP Signaling Pathway in Dentin Development and Diseases. Cells 11: 2216.
Liu Y., Guo W. (2021). SOX factors as cell-state regulators in the mammary gland and breast cancer. Seminars in Cell & Developmental Biology 114: 126-133.
Ma Q., Song C., Yin B., Shi Y., Ye L. (2022). The role of Trithorax family regulating osteogenic and Chondrogenic differentiation in mesenchymal stem cells. Cell Proliferation 55: e13233.
Ming Z., Vining B., Bagheri-Fam S., Harley V. (2022). SOX9 in organogenesis: shared and unique transcriptional functions. Cellular and Molecular Life Sciences 79: 522.
Motyl K. J., Raetz M., Tekalur S. A., Schwartz R. C., McCabe L. R. (2011). CCAAT/enhancer binding protein β-deficiency enhances type 1 diabetic bone phenotype by increasing marrow adiposity and bone resorption. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology 300: R1250-R1260.
Mukherjee S., Luedeke D. M., McCoy L., Iwafuchi M., Zorn A. M. (2022). SOX transcription factors direct TCF-independent WNT/β-catenin responsive transcription to govern cell fate in human pluripotent stem cells. Cell Reports 40: 111247.
Ohba S. (2021). Genome-scale actions of master regulators directing skeletal development. Japanese Dental Science Review 57: 217-223.
Pingault V., Zerad L., Bertani-Torres W., Bondurand N. (2022). SOX10: 20 years of phenotypic plurality and current understanding of its developmental function. Journal of Medical Genetics 59: 105-114.
Kawasaki K., Kawasaki M., Watanabe M., Idrus E., Nagai T., Oommen S., Maeda T., Hagiwara N., Que J., Sharpe P. T., Ohazama A. (2015). Expression of Sox genes in tooth development. The International Journal of Developmental Biology 59: 471-478.
Klum S., Zaouter C., Alekseenko Z., Björklund K., Hagey D. W., Ericson J., Muhr J., Bergsland M. (2018). Sequentially acting SOX proteins orchestrate astrocyte‐ and oligodendrocyte‐specific gene expression. EMBO reports 19: e46635.
Qin X., Jiang Q., Komori H., Sakane C., Fukuyama R., Matsuo Y., Ito K., Miyazaki T., Komori T. (2021). Runt‐related transcription factor‐2 (Runx2) is required for bone matrix protein gene expression in committed osteoblasts in mice . Journal of Bone and Mineral Research 36: 2081-2095.
Reinhold M. I., Naski M. C. (2007). Direct Interactions of Runx2 and Canonical Wnt Signaling Induce FGF18. Journal of Biological Chemistry 282: 3653-3663.
Reznik N., Fass D. (2022). Disulfide bond formation and redox regulation in the Golgi apparatus. FEBS Letters 596: 2859-2872.
Rodríguez-Carballo E., Ulsamer A., Susperregui A. R.G., Manzanares-Céspedes C., Sánchez-García E., Bartrons R., Rosa J. L., Ventura F. (2011). Conserved regulatory motifs in osteogenic gene promoters integrate cooperative effects of canonical Wnt and BMP pathways. Journal of Bone and Mineral Research 26: 718-729.
Schock E. N., LaBonne C. (2020). Sorting Sox: Diverse Roles for Sox Transcription Factors During Neural Crest and Craniofacial Development. Frontiers in Physiology 11: 606889.
Steger J., Cole A. G., Denner A., Lebedeva T., Genikhovich G., Ries A., Reischl R., Taudes E., Lassnig M., Technau U. (2022). Single-cell transcriptomics identifies conserved regulators of neuroglandular lineages. Cell Reports 40: 111370.
Stevanovic M., Drakulic D., Lazic A., Ninkovic D. S., Schwirtlich M., Mojsin M. (2021). SOX Transcription Factors as Important Regulators of Neuronal and Glial Differentiation During Nervous System Development and Adult Neurogenesis. Frontiers in Molecular Neuroscience 14: 654031.
Weider M., Wegner M. (2017). SoxE factors: Transcriptional regulators of neural differentiation and nervous system development. Seminars in Cell & Developmental Biology 63: 35-42.
Wen Q., Jing J., Han X., Feng J., Yuan Y., Ma Y., Chen S., Ho T.V., Chai Y. (2020). Runx2 Regulates Mouse Tooth Root Development Via Activation of WNT Inhibitor NOTUM . Journal of Bone and Mineral Research 35: 2252-2264.
Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., Rahl P. B., Lee T. I., Young R. A. (2013). Master Transcription Factors and Mediator Establish Super-Enhancers at Key Cell Identity Genes. Cell 153: 307-319.
Wu M., Chen G., Li Y.P. (2016). TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Research 4: 16009.
Zhou G., Zheng Q., Engin F., Munivez E., Chen Y., Sebald E., Krakow D., Lee B. (2006). Dominance of SOX9 function over RUNX2 during skeletogenesis. Proceedings of the National Academy of Sciences 103: 19004-19009.