Int. J. Dev. Biol. 66: 163 - 175 (2022)
Special Issue: Developmental Biology in Greece
RNA silencing pathways in plant development and defense
Review | Published: 22 November 2021
Abstract
RNA silencing refers to a conserved eukaryotic process and is regarded as one of the most important processes in plants, with the ability to regulate gene expression both transcriptionally and post-transcriptionally. Different classes of non-coding RNAs (ncRNAs) constitute key components of the RNA silencing pathways and play pivotal roles in modulating various biological processes as well as host-pathogen interactions. One of the most extensively studied classes of ncRNAs are the 20–24 nucleotide (nt) long microRNAs (miRNAs), which are core components of the endogenous gene silencing pathway. miRNAs act as negative regulators of endogenous gene expression through either mRNA-target cleavage, translational inhibition, or DNA methylation, and are inextricably linked to a plethora of developmental processes, such as leaf pattern formation as well as abiotic and biotic stress responses. In this review, we focus on the role of the RNA silencing pathways in the regulation of developmental processes as well as in the plant responses to biotic stress.
Keywords
non-coding RNAs, microRNAs, RNA interference, abiotic stress, biotic stress
Introduction
Plant scientists had acquired confidence in Agrobacterium tumefaciens transformation methodologies in the late 1980s. The door was open for transferring genes of commercial interest into plants. In one of these early attempts, scientists at the Plant Technology Corporation in Oakland, California transformed petunia plants with the aim of intensifying the coloration of the petals of this important ornamental species. The team of Napoli et al., 1990 introduced into the transgenic petunias additional copies of the gene CHALCONE SYNTHASE (CHS), a key gene in the flavonoid pigment biosynthetic pathway. Independently of the above group, another lab in the Netherlands followed a similar strategy (Van Der Krol et al., 1990). Existing knowledge at the time would support an increase in petunia flower coloration. Even ineffectiveness of the additional gene could be attributed to the maintenance of cellular homeostasis. Nevertheless, the scientific community was surprised to see that the introduction of the additional transgene (additional copies of an endogenous gene) led to striking suppression of the coloration. In addition, this suppression (named co-suppression) was shown to be inherent to cuttings. Although the scientists involved realized the significance of their findings, it was only eight years later that we began to understand the mechanisms behind these phenomena. Just a few years after the petunia co-suppression studies, two additional, seemingly unrelated papers, introduced significant new findings into the emerging field of RNA biology. In a seminal 1993 paper in Cell, V. Ambros and colleagues characterized a short ncRNA expressed from the Caenorhabditis elegans (C. elegans) genome abnormal cell-LINeage-4 (lin-4) as a regulator of another gene abnormal cell-LINeage-14 (lin-14) at the post-transcriptional level (Lee et al., 1993). Independently, in the early 1990s, plant virologists were attempting to generate transgenic plants resistant to viruses, through the introduction of genes from the cognate virus into the host plant genome, in an approach reminiscent of vaccination in mammals. In many cases, the resulting transgenic plants were indeed resistant, despite the fact that plants were not known to possess a mammalian-like innate immune system against viruses. In 1994, W. Dougherty, a plant virologist, following a series of findings made by himself and others, presented a hypothesis with regard to the mechanism behind this acquired immunization in plants. He suggested that, since even truncated genes of viral origin were able to confer resistance to the cognate virus, it was possibly small RNAs (sRNAs) derived from the transcript of viral origin that gave the mechanism its sequence specificity (Smith et al., 1994).
It was not until four years later that seminal papers on both C. elegans and plants showed how such a mechanism could function. Following extensive experimentation with C. elegans, Mello and colleagues demonstrated that the key event in triggering RNA-mediated silencing phenomena was the presence of double-stranded RNA (dsRNA) (Fire et al., 1998). Soon afterwards, the authors coined the term RNA-interference (RNAi) to describe this phenomenon. Independently, a few months later, P. Waterhouse and colleagues demonstrated that virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA (Waterhouse et al., 1998). The notion that sRNAs (circa 25 nucleotide, nt, long) are the blueprint of the of RNA silencing mechanism was confirmed in a work published in 1999 by A. Hamilton and D. Baulcombe (Hamilton and Baulcombe, 1999). It was later demonstrated that dsRNAs are cleaved by specialized dsRNA-specific ribonucleases called Dicers into sRNAs that would then serve as mediators of sequence-specific suppression. Although discovered independently in plants and metazoa, Dicer proteins were first identified as the mediators of the cleavage step in the processing of dsRNAs and thus intricately linked to RNAi, several years later (Bernstein et al., 2001). The Dicer group of proteins is well conserved in eukaryotes and is characterized by the presence and order of specific domains, from N- to C- terminal: helicase, Piwi/Argonaute/Zwille (PAZ) domain, 2X Ribonuclease III (RNAse III) and a double-stranded RNA binding domain (dsRBD). The structure of this group of enzymes is characterized by their ability to recognize, bind and cleave dsRNAs, producing small-dsRNA cleavage products, usually in the range of 20-25 nt, hence their name (Ciechanowska et al., 2021). Plants, as exemplified by Arabidopsis thaliana (A. thaliana), typically contain at least four Dicer proteins known as DICER-LIKE (DCL) proteins with specialized roles. Nevertheless, some overlap in DCL function has been described, with the most pronounced being the one between DCL2 and DCL4, which are both involved in anti-viral and RNAi pathways. DCL1 cleaves non-perfect dsRNA substrates and typically produces 21 nt sRNAs involved in post-transcriptional regulation of other genes. This class of sRNAs are known as plant miRNAs (presented in detail below). DCL2 and DCL4 that are involved in the cleavage of fully complementary dsRNAs to produce 22 nt and 21 nt long sRNAs respectively. They are involved in antiviral defense and transgene post-transcriptional suppression known as post-transcriptional gene silencing (PTGS). In addition, DCL4 is also responsible for the generation of a specific group of regulatory sRNAs known as trans-acting sRNAs (tasiRNAs), whereas DCL2 is involved in a phenomenon known as transitivity, allowing the 5’-3’ downstream spread of RNA target cleavage. Finally, DCL3 is responsible for the generation of 24 nt siRNAs, usually as a result of the processing of a specific group of endogenous dsRNA transcripts involved in RNA-directed DNA methylation (RdDM) phenomena (see also below) (Erdmann and Picard, 2020). sRNAs are then taken up by a large group of proteins known as Argonautes (AGOs) that are well conserved in eukaryotes and beyond (recently reviewed by Niaz, 2018). Argonaute proteins are central to all RNA silencing pathways. Generally, they are classified into three groups: true AGOs, PIWI proteins and WAGO proteins. All plant AGOs belong to the first clade, and typically plants possess 10 or more. They have versatile but also partly overlapping functions. In A. thaliana, their functions range from antiviral defense and transgene silencing (typically AGO1,2), to mRNA regulation (AGO1,2,10), tasiRNA production (AGO7) and RdDM (AGO4,6,9) (reviewed by (Borges et al. 2015; Zhang et al. 2015)). AGO proteins, together with the incorporated sRNA, are the effectors of RNA silencing, as part of a larger complex. This complex is usually known as RISC (RNA induced silencing complex) but depending on its function may also have other names. Comprehensive review of proteins involved in the silencing effector complexes such as RISC can be found elsewhere (Zhang et al., 2018b). Plants, fungi and, exceptionally, animals, may also have RNA-DEPENDENT RNA POLYMERASES (RDRs) as important parts of the silencing pathway. In A. thaliana at least three functional RDR proteins have been identified. RDR1 and RDR6 have been characterized as important members of the antiviral and sense-PTGS pathways, while RDR6 has also been implicated in various developmental phenomena such as phase change, and leaf and pistil development (most likely through its role in tasiRNA generation). RDR2 is mainly known for its role in genome maintenance through its function in RdDM (reviewed in Willmann et al., 2011). RDRs and especially RDR6 have been shown to be essential for efficient systemic RNA silencing spread (reviewed in Chen et al. 2018; Mermigka et al. 2014).
The aim of this review is to summarize the role of RNA silencing pathways in response to plant development and defense.
Small non-coding RNAs
For some time, ncRNAs were regarded as transcriptional noise, due to a lack of minimal protein-coding capacity. Later however, there was steady increase in the importance of ncRNAs as transcriptional and post-transcriptional regulators of plant development and defense against both biotic and abiotic stress. At an early stage, small ncRNAs were identified as the blueprint of the RNA silencing pathways, with a significant impact on development and defense (Baulcombe 2004; Hamilton and Baulcombe 1999), and more recently (Hung and Slotkin 2021; Sanan-Mishra et al. 2021). Based on their origin, biogenesis, and mechanism of action, ncRNAs have been categorized as either housekeeping or regulatory ncRNAs (Waititu et al., 2020). Regulatory ncRNAs are further separated into three classes according to their size: short ncRNA (17-3nt), middle-sized ncRNAs (31-200 nt) and long ncRNAs (lncRNA) (> 200 nt). Short ncRNAs includes micro RNAs (miRNA), small interfering RNAs (siRNA), piwi RNAs (piRNA), transcription initiation RNAs (tiRNA), centromere repeat associated small interacting RNAs (crasiRNA) and telomere-specific small RNAs (telsRNA) (Katsarou et al., 2015). Depending on the specific enzymes and RNA silencing pathway used for the mRNA target control, sRNAs could be further divided into endogenous and exogenous sRNAs.
Endogenous gene silencing
Endogenous RNA silencing refers mainly to RNA silencing guided by a class of regulatory sRNAs, the miRNAs. As mentioned above, the first regulatory miRNA, lin-4, was found in C. elegans, controlling the timing of larval development through translational inhibition of its target gene (Lee et al., 1993). Several years later, miRNAs were identified in Drosophila and subsequently in plants (Reinhart et al. 2002; Stark et al. 2003). miRNA biogenesis is triggered by the production of transcripts with extensive fold-back structures, transcribed from endogenous non-coding genes known as MIR genes. Mature miRNAs are non-coding RNA molecules, 20–24 nucleotides in length, without open reading frames or protein coding capacity. miRNAs are negative regulators of endogenous gene expression either through mRNA-target cleavage, translational inhibition or DNA methylation (Song et al., 2019). miRNAs have been associated, among various physiological processes, with the control of plant development, growth, hormone homeostasis, as well as biotic and abiotic stress adaptation, by regulating the expression of many transcription factors and stress-responsive proteins (Li et al., 2017).
The microRNA pathway
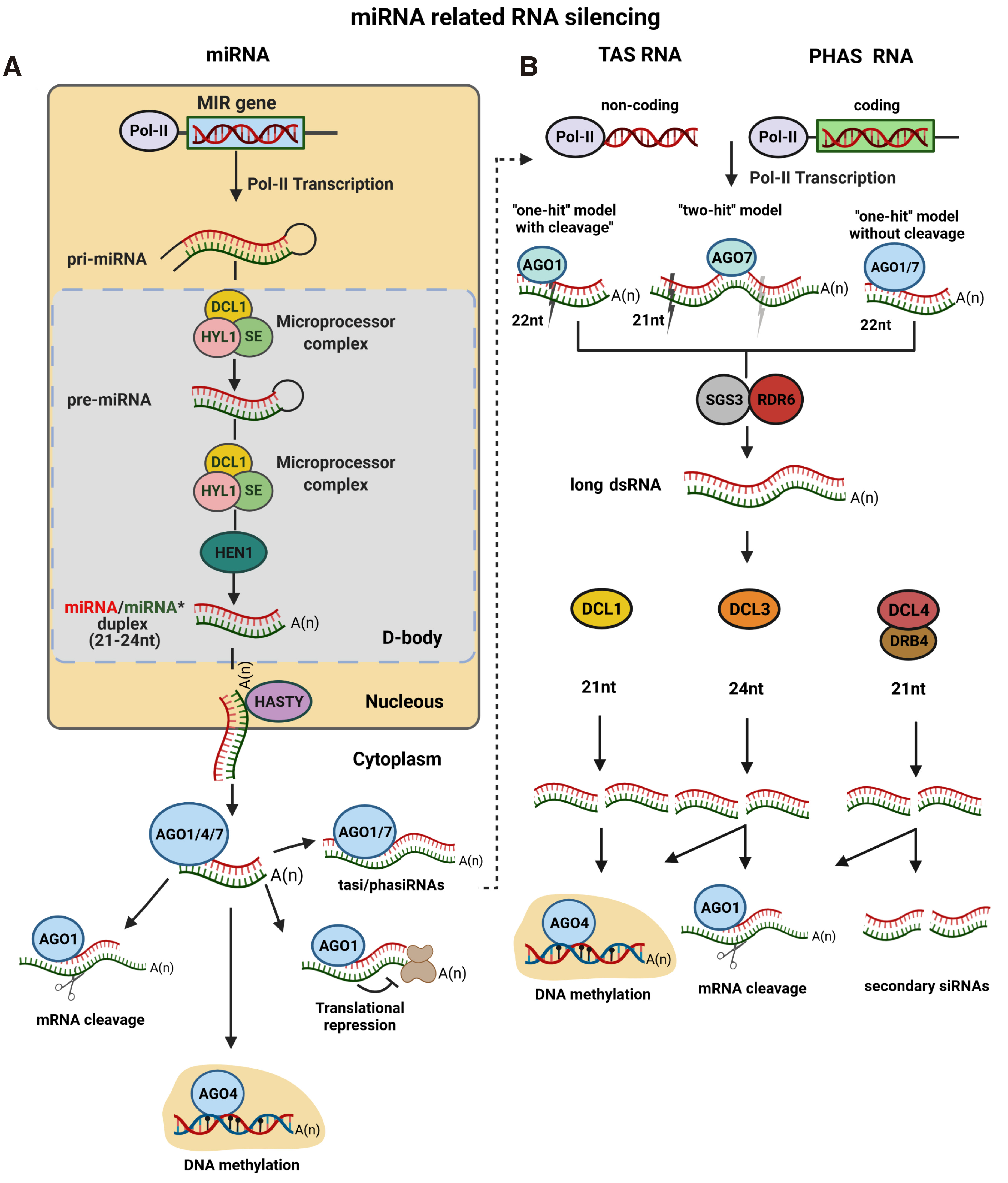
The process of miRNA biogenesis consists of four distinct steps (described below). The majority of miRNAs are generated from the transcription of MIR genes located in intergenic regions, although several miRNA precursors have also been identified in introns and promoter regions of Brassicaceae (Barciszewska-Pacak et al. 2016; Megraw et al. 2006; Yan et al. 2012). MIR genes are transcribed by DNA-DEPENDENT RNA POLYMERASE II (Pol-Il) to give rise to long, single-stranded, 5’ capped and 3’ polyadenylated primary miRNAs (pri-miRNAs). The imperfect stem-loop structure of the pri-miRNAs is then recognized by the RNase III enzyme DCL1 and processed into short precursor RNAs (pre-miRNAs). In this step, the dsRNA-binding protein HYPONASTIC LEAVES 1 (HYL1) and the zinc-finger protein SERRATE (SE), which along with DCL1 constitute the Microprocessor complex, are also involved. Pre-mRNAs are further processed by the Microprocessor complex to generate imperfect miRNA/miRNA* duplexes, consisting of 2-nt 3' overhangs at each end (Wang et al., 2019). MIR biogenesis from gene transcription to generation of the mature miRNA/miRNA* complex takes place in the nucleus and more specifically in distinct nuclear dicing–bodies (D-bodies). The miRNAs are then stabilized by the addition of a methyl-group to the 2’-hydroxylgroup of the 3’ terminal nucleotide overhangs by the methylase HUA ENHANCER 1 (HEN1), while it is transferred to the cytoplasm with the aid of the HASTY protein (Stepien et al., 2017). One strand of miRNA/miRNA* duplex, usually the miRNA strand, is then selected as a guide strand and loaded onto AGO proteins, mostly AGO1, but also in some cases by either AGO2, AGO4, AGO7 or AGO10, thus forming the RISC that leads to translation inhibition, cleavage of the complementary target mRNA or DNA methylation in a sequence-specific manner (Gao et al. 2021; Song et al. 2019) (Fig. 1A).
Fig. 1. Schematic representation of the biogenesis and function of miRNAs and TAS/PHAS mRNAs.
(A) Schematic representation of the biogenesis of miRNAs. MIR genes are initially transcribed by Pol-II to ssRNA that forms an internal fold-back loop, the pri-miRNA. pri-miRNA is then processed by the Microprocessor complex to produce pre-miRNA, which is subjected to a second round of processing by the Microprocessor complex to form the miRNA/miRNA* duplex. This is then methylated by HEN1 and transported to the cytoplasm by HASTY. The miRNA guide strand is selected as a guide strand and loaded onto AGO1 proteins to form the RISC, which leads to translation inhibition, cleavage of the complementary target mRNA or DNA methylation in a sequence-specific manner. The nucleus area is denoted in a beige frame. (B) Schematic representation of the biogenesis of tasiRNAs and phasiRNAs. The tasiRNA and phasiRNA biogenesis starts when mRNA targets are cleaved by a miRNA precursor. If the cleavable miRNA is 22 nt long, it follows the ‘one-hit’ model where the miRNA has a single target site and slices the target via AGO1. When the cleavable miRNA is 21 nt long, it follows the ‘’two-hit’’ model, according to which the 21 nt trigger miRNA has two target sites, but only one can be cleaved by AGO7. An alternative ‘’one-hit’’ model has recently been proposed, where the fragments produced by the miRNA-AGO complex induce the production of secondary siRNA by AGO1 or AGO7, even in the absence of cleavage events. In all cases, the miRNA cleavage products are stabilized by SGS3 and converted into a dsRNA by RDR6. The dsRNAs are then processed, mainly by DCL4/DRB4, and result in the generation of 21 nt siRNAs that are recruited by the AGO1 complex to mediate the cleavage of target mRNA or induce the production of secondary siRNAs. Processing, possibly by DCL1, leads to the generation of 21 nt siRNAs that are incorporated into AGO4 protein to mediate the methylation at their own DNA loci. An alternative process by DCL3 results in the production of 24 nt siRNAs that may associate with AGO1 to cleave target sequences or with AGO4 for DNA methylation. (Created with BioRender.com).
Microprocessor complex
DCL1 is transcribed from the maternally inherited allele and its activity is essential for plant development, as complete loss of function mutations of dcl1 cause embryo lethality in A. thaliana (Ray et al., 1996). DCL1 constitutes the major factor in the plant miRNA biogenesis pathway and produces 21 nt sRNAs from the processing of pri- and pre-miRNA transcripts. DCL1 co-localizes with D-bodies where it interacts with Microprocessor complex partners HYL1 and SE, which contribute to its processing accuracy and efficiency (Dong et al., 2008). DCL1 is important for many developmental processes, including the transition from the juvenile to the mature phases of plant development. Accordingly, even a small reduction of DCL1 levels leads to various developmental defects in both A. thaliana and N. benthamiana plants (Katsarou et al. 2018; Schauer et al. 2002). In addition, DCL1 has been associated with DNA methylation, contributing to the silencing of certain transposons and facilitating the biogenesis of DNA virus siRNAs by other DCLs (Ciechanowska et al., 2021).
SE constitutes the plant orthologue of the mammalian gene Arsenite resistance 2 (ARS2) that encodes a C2H2–type zinc finger protein (Rédei, and Hirono, 1964) and promotes the recognition and cleavage of pri- and pre-miRNAs. SE mutants in A. thaliana, as well as in Nicotiana tabacum (N. tabacum) and N. benthamiana plants, show decreased mature miRNA levels, implicating their role in miRNA biogenesis (Kryovrysanaki et al. 2019; Lobbes et al. 2006). SE has the ability to interact with many factors that enhance its role in RNA processing pathways. It has been shown that SE interacts with both subunits of CAP-BINDING PROTEINS (CBP20 and CBP80), thus acting as a bridge connecting the miRNA pathway and the spliceosome (Raczynska et al., 2014). This interaction also promotes association with Ser5- and Ser2-phosphorylated RNA Pol-II complexes, regulating the transcription of mainly intronless genes (Speth et al., 2018). In another pathway, SE recruits and interacts with the nuclear exosome targeting (NEXT) complex to promote pri-miRNA degradation (Bajczyk et al., 2020), while it can also inhibit the processing of pri-miRNAs by inducing their structural alteration via interaction with CHROMATIN REMODELLING FACTOR 2 (CHR2) (Wang et al., 2018).
HYL1 is another core component of the miRNA biogenesis pathway. HYL1 promotes accuracy in DCL1 processing through its involvement in the correct loading of DCL1 onto its substrate to initiate pri-miRNA processing assisted by its two RNA binding sites and its ability to form homodimers (Dong et al., 2008). In addition, HYL1 may also protect pri-miRNAs from nuclear exosome attack and stabilize them (Gao et al., 2020). Moreover, it has recently been shown that HYL1 localizes to polysomes on the endoplasmic reticulum (ER), where it associates with AGO1 and ALTERED MERISTEM PROGRAM 1 (AMP1), monitoring the distribution of AGO1 onto polysomes and repressing the translation of target genes when binding to their mRNA. It has been demonstrated that HYL1 binds to the miRNAs and mRNAs of miRNA-targeted genes to form a miRNA-mediated effector complex leading to translational repression of the mRNA of target genes (Yang et al., 2021).
miRNAs and development
Due to their sessile nature, plants must utilize cost-effective pathways for the regulation of growth and development, both under normal growth conditions as well as under adverse environmental changes or threats from various biotic factors. To achieve this regulation, both sRNAs and miRNAs, including inducible miRNA-guided sRNAs, as well as exogenous siRNAs, are utilized. It has been shown that evolutionarily conserved miRNAs usually have conserved gene targets, while in some cases a single miRNA can target multiple members of a gene family. miRNAs regulate developmental processes such as leaf morphogenesis, vegetative phase change, flowering time and responses to environmental cues, and control gene expression through cleavage of their targets, translational repression and host genome DNA methylation (Li and Zhang 2016; Yu et al. 2018). Although translational repression was initially considered to be uncommon in plants, there are now numerous cases where crucial regulation of plant development genes has been demonstrated at the protein level. Two examples are the regulation of APETALA2 (AP2) and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3) by miR172 and miR156/7, respectively, which are involved in juvenile to adult phase transition (Yu et al., 2018). In both cases, abnormal miRNA levels lead to changes in the levels of protein production, while transcript levels were comparable. Similar observations have been made for other miRNAs, including miR159, miR171, miR395, miR398 and miR834 miR164, miR165/6 (Yu et al., 2018).
In addition, there have been reported cases of miRNAs that share common target genes important for plant growth. To date, miR165 and miR166 are involved in shoot apical meristem (SAM)-related development, including leaf polarity, by repressing the expression of HOMEODOMAIN LEUCINE ZIPPER III (HD-ZIP III). This occurs via an antagonistic interaction between AGO1 and AGO10, with AGO10 acting as a decoy for miR166/165 to maintain the SAM, preventing their incorporation into AGO1 complexes and the subsequent repression of HD-ZIP gene expression (Li and Zhang, 2016).
Regulation and control of specific pathways during plant growth/development is usually determined by more than one miRNA, with distinct miRNAs expressed in specific tissues at a given time. Examples of such interactions between miRNAs include the interplay of miR160, miR165/166 and miR394 at leaf initiation, or the networking of miR165/166, miR390 and miR396 in the control of leaf polarity. Different examples are the interplay of miR156, miR159 and miR172, which determines the phase transition from juvenile to adult and from vegetative to reproductive phase, as well as the interplay of miR164, miR319 and miR390, which contributes to leaf senescence (Yang et al., 2018).
miRNAs in abiotic stress responses and plant homeostasis
Plants have developed the ability to use miRNA regulated gene transcription in order to respond to environmental cues and stress conditions such as extreme temperature, light conditions, nutrient deficiency and salt stress. The molecular and hormonal regulatory mechanism behind these responses have been extensively described elsewhere (Ali et al. 2020; Song et al. 2019; Waititu et al. 2020). Different environmental and stress conditions induce the expression of distinct miRNAs, while in some cases differential miRNA expression (induction versus suppression) can facilitate a distinct plant response to stress conditions or facilitate adaptation to changes in the environment (Sunkar et al. 2012; Waititu et al. 2020). Several well-characterized miRNAs respond broadly to various abiotic stresses, while in other examples, miRNAs respond only under specific conditions. miR159, miR160 and miR167 are conserved miRNAs that negatively regulate abscisic acid (ABA) and AUXIN RESPONSE FACTORS (ARFs), affecting a plethora of plant characteristics such as phyllotaxis, lateral root formation, hypocotyl length and fertility (Bhogireddy et al., 2021). In A. thaliana, miR398 is inhibited by various stresses, including oxidative stress, salt tolerance, ABA, and high copper, while miR169a and miR169c are both inhibited by drought and ABA stress. In contrast, the expression of miR393 was induced in A. thaliana by salt, drought, cold, ABA, and UV-B stresses. Furthermore, in some cases, a single miRNA has been shown to respond differently depending on the type of abiotic stress. For example, A. thaliana miR169 was induced by salt, cold, and UVB irradiation but suppressed by drought, heat and ABA treatment (Waititu et al., 2020).
Under certain conditions, a whole network of miRNAs can be altered in order for the plants to adjust and survive. This network of miRNAs could be the same in different plant species, or may differ between species. For example, in both A. thaliana and maize, miR169, miR171, miR395, miR397, miR398, miR399, miR408, and miR827 were all up-regulated in response to nitrogen deficiency. On the contrary, miR168, miR188, and miR397 were all induced in A. thaliana in response to drought stress, while in maize the same miRNAs were suppressed. (Song et al. 2019; Waititu et al. 2020; Yan et al. 2016).
Classes of endogenous siRNAs
In addition to miRNAs, plants also produce various classes of endogenous sRNAs that are categorized according to their biogenesis processes and the enzymes with which they interact. Described below are the different classes of miRNA-triggered secondary siRNAs that influence plant defense and development.
Phased siRNAs and Trans-acting siRNAs
Phased siRNAS (phasiRNAs; usually 22 nt in length) and trans-acting RNAs (tasiRNAs; usually 21 or 24 nt in length) are two classes of miRNA-triggered secondary siRNAs that are the cleavage products of mRNA transcripts targeted by miRNAs. The precursors of tasiRNAs are transcribed from non-coding genes, TAS genes, while the precursors of phasiRNAs are transcribed from protein-coding genes, PHAS genes. The biogenesis of tasiRNAs, and in most cases that of phasiRNAs, is identical. The miRNA cleavage products are stabilized by SUPPRESSOR OF GENE SILENCING 3 (SGS3) and converted into a dsRNA form by RDR6. dsRNAs intermediates are then processed, mainly by DCL4 and DOUBLE-STRANDED RNA BINDING FACTOR4 (DRB4), and are finally recruited to AGO1 complex to mediate the cleavage of target mRNA or generate secondary phasiRNAs 21 nt long, (Allen et al. 2005; Sanan-Mishra et al. 2021). Alternatively, processing of dsRNAs, possibly by DCL1, leads to the generation of 21 nt siRNAs that are incorporated into AGO4 protein to mediate the methylation at their own DNA loci. While, in a different case, processing by DCL3 results in the production of 24 nt siRNAs that may associate with AGO1 to cleave target sequences or with AGO4 for DNA methylation. Two main mechanisms have been described by which the final secondary tasiRNAs and phasiRNAs are generated. The ''one hit'' model in which, usually, a 22 nt miRNA has a single target site and slices the target via AGO1, and the ‘’two-hit’’ model where a 21 nt trigger miRNA has two target sites, only one of which can be cleaved even though both interact with the miRNA via AGO7. Moreover, an alternative pathway has been proposed, according to which there is no need for a cleavage on the miRNA-target upon loading to AGO1 or AGO7. Typically, tasiRNAs guided by AGO1 and AGO7 lead to the silencing of endogenous loci in trans, regulating gene expression and various developmental processes, while phasiRNAs lead to cleavage of their targets (Deng et al. 2018; Fei et al. 2013; Liu et al. 2020). For detailed description of the tasi/phasiRNA biogenesis and their function, see Fig. 1B (Deng et al. 2018; Liu et al. 2020).
Plant phasiRNAs are known to function in diverse biological processes. One of the best characterized PHAS loci identified to date is the miR390-AGO7 complex. This PHAS locus associates with the TAS3 and targets ARF family transcription factors involved in developmental transitions, embryo development, root structure, SAM development, leaf morphology and flower and phytohormone cross-talk (Allen et al. 2005; Liu et al. 2020). PHAS loci within protein-coding genes encode larger subgroups of phasiRNAs including, among others, NUCLEOTIDE-BINDING LEUCINE-RICH REPEAT (NLR), PENTATRICOPEPTIDE REPEAT (PPR), as well as MYB transcription factor (TF) loci. These loci, and the phasiRNAs produced, function as negative regulators in many biological processes, such as disease resistance, plant vegetative and reproductive development, seed germination, and plant parasitism (Fig. 1B) (Liu et al., 2020).
Heterochromatic siRNAs
Heterochromatic siRNAs (hcsiRNAs) are 23-24 nt in length, generated from the transcription of intergenic or repetitive regions of the plant genome by Pol-IV and also possibly by Pol-V. RDR2 is responsible for the amplification of the dsRNA, which in turn is processed by DCL3 to produce the mature hcsiRNAs. At this stage, siRNAs are incorporated into the RISC complex by the AGO4 activity, leading to transcriptional gene silencing (TGS) by guiding the methylation of DNA and histones through the RdDM pathway. Depending on the sequence similarity of the DNA targets, hcsiRNAs can act also in trans (Matzke and Mosher, 2014).
Natural antisense transcript-derived siRNAs
Natural antisense transcripts (NAT)-derived siRNAs (nat-siRNAs) are formed by base-pairing of complementary transcripts. When the siRNAs are generated from the transcription of both strands of the same genomic loci they are termed cis-NAT, while, when the transcribed product derives from complementary DNA sequences of different loci, they are known as trans-NAT (Borges et al., 2015). Although there is currently little information available, nat-siRNAs have been associated with stress alleviation, including salt stress tolerance, Pseudomonas syringae (P. syringae) pv. tomato DC3000 resistance and hormone synthesis regulation (Sanan-Mishra et al., 2021).
Epigenetic modifications in plants
Plants, being sessile organisms, have evolved specific gene regulatory mechanisms to ensure their survival in changing environmental conditions and during plant development. Epigenetic regulation consists of covalent changes of DNA and histones that influence transcriptional activity of chromatin without changing the DNA sequence. DNA methylation, sRNAs and histone modifications are the three epigenetic elements in plants that have received the most attention. DNA methylation and chromatin modification can be triggered by sRNAs (Thiebaut et al., 2019). Endogenous RNA-directed gene silencing can take place at the post-transcriptional or the transcriptional level. PTGS occurs after the formation of mRNA, when microRNAs (21–23 nt in length) mark gene transcripts with homologous sequence and direct their degradation or block their translation. TGS takes place in the nucleus where sRNAs drive epigenetic modifications in a sequence-specific manner (Matzke and Mosher, 2014).
The RdDM pathway
TGS via RdDM directs the epigenetic regulation of genome stability, gene expression, as well as a general regulation of the genome in response to growth, development and stress signals. DNA methylation is a conserved process from plants to mammals and is crucial for development. DNA methylation takes place at the 5’ carbon of the cytosine ring as a result of activity by several enzymes. In contrast to mammals, plant DNA methylation can take place in all cytosine sequence contexts (i.e. CG, CHG and CHH, where H represents A, T or C) (Li and Zhang 2014; Zhang et al. 2018a). The RdDM pathway is crucial for de novo methylation in plants and has been described as two different pathways (canonical and non-canonical). These can be further separated into two parts, namely the production of sRNAs and the methylation of the target DNA loci.
The canonical RdDM pathway starts with the transcription of CLASSY (CLSY) proteins and SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) from RNA Pol-IV to produce a single-stranded RNA (ssRNA) that is around 30 to 45 nt long. These ssRNAs are then processed by RDR2, and dsRNAs are generated. The dsRNAs are then cleaved into 24 nt siRNAs by DCL3 and are stabilized by methylation at the 3’-OH group by HEN1.
In the second part of the pathway, AGO proteins load one strand of the 24 nt siRNAs to form the AGO-sRNA duplex. Only AGO4, AGO6, and AGO9 are able to load 24 nt siRNAs, with AGO4 being the main AGO protein utilized in the canonical RdDM pathway. The AGO-sRNA duplex is then able to recognize and bind complementary RNA sequences. These RNA complementary sequences are non-coding transcripts that are produced by RNA Pol-V and act as scaffolds with which the 24 nt siRNAs base-pair. The access of Pol-V to its target sites is facilitated by the DEFECTIVE IN RNA-DIRECTED DNA METHYLATION (DRD1) complex, named after the three components DEFECTIVE IN RNA-DIRECTED DNA METHYLATION (DRD1), DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) and RNA-DIRECTED DNA METHYLATION (RDM1) that possibly unwind the DNA downstream of Pol-V. Pol-V then recruits AGO4 by interacting with the NUCLEAR RNA POLYMERASE D1B (NRPE1), the largest subunit of Pol-V, and the SUPPRESSOR OF TY INSERTION 5-LIKE (SPT5L) through an AGO hook motif that is common to both. All these lead to the recruitment of the DNA methyltransferase enzyme DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), which locally catalyzes de novo DNA methylation in a sequence-independent manner (Erdmann and Picard, 2020).
The non-canonical RdDM pathway shares common features with the canonical pathway and acts as a link between PTGS and RdDM. It is differentiated in the first part of the pathway as it uses alternative entry points from which small RNAs can be induced to the canonical RdDM, while the second part of the pathway utilizing Pol-V and DRM2 remains the same for both types of RdDM.
There are at least six non-canonical RdDM pathways, such as DNA methylation, directed by inverted repeats and miRNAs, RDR6 RdDM, RDR6-DCL3 RdDM, Pol-IV-NERD RdDM, double strand breaks that recruit RdDM-related proteins and dicer independent RdDM. All the above mechanisms have been found to influence PTGS and whole genome DNA methylation patterns (Cuerda-Gil and Slotkin, 2016).
Role of RdDM in plant growth and development
RdDM is crucial for many biological functions such as silencing of transposable elements (TEs), genome stability, short and long-range silencing and stress responses, as well as development and reproduction (Erdmann and Picard, 2020). During plant growth and development, DNA methylation levels in different cell types and tissues is strictly controlled to prevent abnormal growth. However, DNA methylation levels within the promoter regions of homologous genes can dramatically differ between model plants such as A. thaliana and crop species. Only 5% of promoter regions in A. thaliana are methylated, suggesting that mutations affecting DNA methylation may not dramatically affect growth and development. In contrast, crop plants with much larger genomes and containing a high number of TEs near to coding sequences are prone to DNA methylation of promoter regions, leading to severe growth and developmental abnormalities (Zhang et al., 2018a). This is the case for ago9 mutants of maize that fail to complete meiosis and generate functional diploid gametes, whereas ago9 mutants of A. thaliana do not show fertility defects. These different developmental outcomes could be explained by the effect these mutations have in the suppression of TEs that are close to different developmental regulators in each species (Matzke and Mosher, 2014).
RdDM is also important for plant reproduction, as it is involved in gamete formation and seed development. The support cell undergoes epigenetic reprogramming and loss of all its epigenetic marks, re-activating TEs that were previously silenced. sRNAs of these TEs trigger RdDM in the support cell and the sRNAs are then moved from the support cell to the germ line to reinforce TE silencing in the next generation. Roots may experience a similar process to sustain TE silencing in stem cell populations. RdDM is also involved in the regulation of imprinting expression of certain genes in the endosperm during seed development in flowering plants, as well as in mediating the gene dosage effects in seeds derived from interploid crosses. FLOWERING WAGENINGEN (FWA) gene, which is responsible for proper flowering time in A. thaliana, is mainly suppressed due to hypermethylation at tandem repeats at its promoter region by RdDM. Loss of methylation results in a late-flowering phenotype, which is heritable to next generation and acts as an epi-allele (Erdmann and Picard, 2020).
Ripening in non-climactic fruits is another aspect of plant development recently shown to be regulated by RdDM. In climacteric fruits, such as tomato, hypomethylation of genes involved in fruit ripening generally results from the increased expression of DNA demethylase genes. However, in strawberries, a non-climacteric fruit, changes in the expression of genes involved in fruit ripening is not associated with an increased expression of DNA demethylases. Instead, it is suggested that during strawberry ripening, the broad downregulation of several components of the RdDM pathway results in the hypomethylation of ripening genes (Cheng et al., 2018).
RNA silencing in defense
The RNA silencing pathway targeting exogenous sequences, such as transgenes and viruses, also known as PTGS or RNAi, uses the sequence-specific recognition of foreign sequences to target them for degradation (Hung and Slotkin 2021; Kalantidis et al. 2008). The key components of this pathway involve DCL2 to 4 generating 22, 24 and 21 nt sRNAs respectively, associating with AGO1 (21/22 nt siRNAs) or AGO4 (24 nt siRNAs) for either target-sequence degradation, the generation of secondary siRNAs, or DNA methylation (Dalakouras et al., 2020). This silencing pathway is non-cell autonomous, with the initiation of RNAi in a cell moving both locally and systemically throughout plants to protect against subsequent viral entry into other host cells (Kalantidis et al. 2008; Zhang et al. 2019). This spread is facilitated by several key host proteins, including RDR6 for extensive local spread, as well as the association of RDR6 with the 22nt/AGO1 pathway, which results in transitivity, or the spread of the silencing signal along the target sequence, leading to amplification of the silencing signal, a key feature of plant defense against viruses (Dalakouras et al. 2020; Mermigka et al. 2014; Zhang et al. 2019).
Silencing against viruses
Viruses are obligate parasites and rely on host plants for replication, movement and transmission. Viral-encoded factors in combination with host-encoded factors are necessary for this interaction. Host plants provide many of the necessary requirements for completion of the viral life cycle, sometimes referred to as susceptibility factors, while also possessing a range of defense pathways to protect against virus infection (Garcia-Ruiz, 2019). Interactions between plants and viruses are governed by the presence or absence of factors encoded by both the host and pathogen, resulting in either compatible or incompatible reactions, also known as susceptibility or resistance, respectively. Some of these factors have a critical effect on plant development, influencing the outcome of the host plant-virus interaction. One of these is RNA silencing.
Plant viruses exploit a great diversity of genome types, including DNA or RNA, which utilize a range of replication strategies. RNA viruses encode RNA-dependent RNA polymerases and replicate in the cytoplasm, the single-stranded DNA (ssDNA) viruses replicate in the nucleus, and the reverse-transcribing double-stranded DNA (dsDNA) viruses encode a reverse-transcriptase and replicate their nuclear-expressed greater-than-genome-length RNAs into genomic DNA copies in the cytoplasm. The detection of viral-derived dsRNA replication intermediates, or self-complementary regions of RNA sequences, triggers the antiviral RNAi pathway to produce virus-derived siRNAs, targeting the viral mRNAs as well as genomic RNAs (Ding and Voinnet 2007; Kalantidis et al. 2002; Pantaleo 2011). For RNA viruses, the dominant players in this pathway are 21 nt siRNAs generated by DCL4, and to a lesser extent, 22 nt siRNAs generated by DCL2.
In contrast, for DNA viruses, the genomic DNA must enter the nucleus for expression. Association of the viral DNA with host machinery utilizes host RNA Pol-II for transcription of viral RNAs. DNA viruses must therefore avoid host RdDM directed by 24 nt siRNAs in conjunction with DCL3, as well as the RNA-directed silencing pathways (Pooggin, 2013). For geminiviruses, their bi-directional genome expression strategy results in the presence of overlapping, self-complementary sequences that are a trigger of RNAi. To avoid RdDM, a range of strategies are employed, including suppression of cytosine methylation and transcriptional silencing by several proteins encoded by either viral genomic or satellite DNAs, together with the rescue of viral DNA from methylation by Rep-mediated replication (Pooggin, 2013). In members of the Caulimoviridae family, multiple transcripts are produced from the viral DNA in the nucleus, including the large 35S transcript, which acts as a polycistronic mRNA for translation of viral proteins and is derived from covalently-closed circular DNA, together with an aberrant small non-coding 8S RNA generated from unrepaired open-circular viral DNA upon nuclear entry. The latter initiates the generation of large numbers of virus-derived small interfering RNAs (vsiRNAs), which act as a decoy by engaging all four DCLs, together with their associated AGOs (Pooggin, 2013). In addition, P6/TAV in Cauliflower mosaic virus (CaMV) acts as a VSR by interfering with the secondary amplification of siRNAs (Pooggin 2013; Pooggin and Ryabova 2018). Avoidance of the RdDM pathway should be managed by the alternating nuclear/cytoplasmic phases of viral expression and replication (Pooggin, 2013).
Involvement of antiviral siRNAs in plant development
In addition to direct interaction of the plant siRNA pathway targeting viral RNAs, antiviral RNA silencing pathways play a distinct role in plant development (Jin et al., 2020). The generation of vsiRNAs with similarity to host genes can have a direct effect on viral pathogenicity (Huang et al., 2016). Several studies have demonstrated that vsiRNAs have a direct effect on plant growth, including developmental defects often associated with symptoms of viral infection. The Y-satellite of Cucumber mosaic virus (CMV) contains a 22 nt sequence complementary to the tobacco CHLI gene involved in chlorophyll biosynthesis. Presence of the Y-satellite initiates siRNAs targeting CHLI, resulting in down-regulation of CHLI expression, leading to the severe yellowing symptoms associated with the presence of Y-satellite (Smith et al., 2011). Similarly, in barley plants infected with Barley yellow dwarf virus-GAV, (BYDV-GAV) a vsiRNA targeting chlorophyll synthase results in yellowing symptoms, while in wheat plants a vsiRNA generated from Chinese wheat mosaic virus (CWMV) regulates expression of a vacuolar (H+)-PPase to inhibit cell death and maintain a weak alkaline environment in the cytoplasm, which enhances CWMV infections (Shen et al. 2020; Yang et al. 2020). Stunting and leaf-curling in tomato was associated with a vsiRNA derived from a 25-nt intergenic region sequence of Tomato yellow leaf curl virus (TYLCV), which induces silencing of the long non-coding RNA SlLNR1 in susceptible but non-resistant plants (Yang et al., 2019). In some cases, widespread silencing of host genes has been associated with the presence of vsiRNAs following infection, while differential expression of several silencing component genes crucial to plant antiviral defense was observed in rice infected with Southern rice black-streaked dwarf virus (SRBSDV) (Xu and Zhou, 2017).
Virus-derived miRNAs/host gene expression
In addition to their roles in plant growth and development, signal transduction, protein degradation, and response to biotic and abiotic stresses, miRNAs also play critical roles in plant-virus interactions. In antiviral defense, two main roles of miRNAs are indicated, either directly through targeting of viral RNAs, or indirectly through the biogenesis of siRNAs that are involved in the antiviral response (Liu et al., 2017).
Viral infection can directly or indirectly affect plant miRNA expression, with increased levels of some miRNAs associated with higher levels of virus, suggesting they have a pro-viral activity, while in other examples a decrease in miRNA expression can also be beneficial for virus infection, suggesting that down-regulation of miRNAs with antiviral activity can benefit viral infection. In some cases, the levels of a miRNA can fluctuate over the course of virus infection. Some viruses interact with specific miRNAs that may not be targets of all viruses, and in some cases even strain-specific differences in miRNA targeting has been observed, while the effect of a virus on host miRNAs may differ between host plants or tissues (Yin et al., 2014). A number of studies have also shown that virus infection is associated with the presence of novel miRNAs that are not present in the absence of virus infection. Many of the miRNAs responsive to viral infection are involved in plant development and defense, and probably contribute to symptoms development as well as influencing the outcome of infection (Liu et al., 2017).
To combat the plant RNA silencing system, viruses have evolved a large and varied class of proteins known as viral suppressors of RNAi, or VSRs. These are usually multifunctional proteins with additional key activities in the viral life cycle, including movement, replication and encapsidation. They manage to do this either by direct binding of RNAs or plant proteins, or indirectly, through regulation of other host factors such as miRNAs, which regulate members of the silencing pathways (Jin et al., 2020). In some cases, specific binding of miRNAs by VSRs impedes their normal function, while selective non-binding of alternative miRNAs has a direct effect on plant growth and defense. This was recently demonstrated for tombusvirus p19 and cucumovirus 2b VSRs, which differentially bind key miRNAs related to AGO1/RISC activities contributing to the regulation of plant defense gene expression in favor of virus infection (Pertermann et al., 2018). Alternatively, other viral proteins may also influence miRNA biogenesis. In Rice stripe virus (RSV), the non-structural NS3 protein interacts with OsDRB1, a component of the miRNA biogenesis pathway, and induces the accumulation of miRNAs targeting genes involved in pathogen resistance, which enhances viral infection (Zheng et al., 2017).
Finally, in several cases there is now evidence that plant virus genomes can encode functional miRNA sequences. This was first demonstrated with Hibiscus chlorotic ringspot virus (HCRSV), which was predicted to encode 5 vir-miRNAs, one of which was confirmed experimentally (Gao et al., 2013). Similar results have been found in Sugarcane mosaic virus (SCMV) and Banana bract mosaic virus (BBrMV) with a range of host targets predicted in each case (Sankaranarayanan et al. 2020; Viswanathan et al. 2014).
RNA silencing and viroids
Viroids are the smallest known pathogens and are restricted to plant hosts. They are non-encapsidated, non-coding, circular ssRNAs 250-400 nucleotides long, classified into two major families, Pospiviroidae and Avsunviroidae. Viroids of the former replicate in the nucleus, and those of the latter in the chloroplasts. Potato spindle tuber viroid (PSTVd) is the type species of pospiviroids and replicates in the nucleus via an asymmetric rolling circle mechanism, transcribed by RNA Pol-II producing dsRNAs. (Rao and Kalantidis 2015; Wang et al. 2004). In N. benthamiana, all four DCLs seem to be involved in the production of viroid-derived siRNAs (vd-siRNAs) (Dadami et al. 2013; Katsarou et al. 2016). Transcripts of PSTVd and peach latent mosaic viroid (PLMVd) are processed into vd-siRNAs when incubated with DCL-containing in vitro extracts (Itaya et al., 2007). The involvement of RDRs in the processing of viroid RNAs is not clear, except for RDR6, which has been linked with the PSTVd exclusion from the meristem (Mermigka et al., 2014). In any case, vd-siRNAs are loaded onto AGOs to target the viroid and the replication intermediates. DCL4-derived 21 nt siRNAs are usually loaded onto AGO1, leading to the assembly of RISC that recognizes and cleaves complementary transcripts (Minoia et al., 2014). DCL2-derived 22 nt siRNAs are also loaded onto AGO1; in this case, however, instead of RNA degradation, the complex induces either secondary siRNA formation via RDR6 or translation inhibition of the target RNA (Wu et al., 2020). DCL3-derived 24 nt siRNAs are mainly loaded onto AGO4 and are involved in RdDM. Viroid siRNAs are both 5’-phosphorylated and 3’-methylated, of both polarities, and with a preference for specific regions of the viroid RNA (Wassenegger and Dalakouras, 2021). It is important to note that vd-siRNAs trigger PTGS of homologous transcripts in the cytoplasm, but not in the nucleus (Dalakouras et al., 2015). Importantly, the mature viroid RNA is resistant to degradation by RISC, probably because of its secondary structure (Itaya et al., 2007). Tomato plants expressing a hairpin PSTVd transgene were resistant to viroid infection, suggesting that RNAi targets RNA molecules essential for PSTVd infection (Schwind et al., 2009).
Viroids and the miRNA pathway
Due to the functional and structural similarities between viroids and miRNAs, it has been suggested that viroid RNAs could act as miRNA-like precursors, leading to the production of miRNA-like vd-siRNAs, which could target host genes (Wang et al., 2004). This suggestion is supported by the finding that satellite RNAs, which share similarities with viroid RNAs, can act as miRNAs to suppress host genes. To assess the involvement of DCL1 in viroid infection, DCL1 knockdown tobacco plants were generated and PSTVd titers were compared to those of wild type plants. The results showed a less efficient infection in DCL1 knockdown plants (Dadami et al., 2013). However, DCL1 downregulation was only moderate, probably due to embryo-lethality of DCL1 knockout (Qin et al., 2017). A more recent study showed that in both SE and DCL1 knockdown tobacco plants there is a strong reduction of pospiviroid titers, with SE also involved in the miRNA pathway. These findings suggest a role of SE and DCL1 in viroid infectivity, and thus the interaction of viroids with components of the miRNA pathway (Dadami et al. 2013; Katsarou et al. 2016; Kryovrysanaki et al. 2019).
Viroid siRNAs and host gene modulation and development
Viroids can strongly induce and be targeted by RNA silencing, due to the formation of dsRNAs during their replication (Pallás et al., 2012). Given the large amount of viroid sRNAs accumulating during viroid infections, it has been suggested that they potentially target host mRNAs, leading to the development of disease symptoms (Tsushima et al., 2014). Early evidence supporting the above assumption was the development of characteristic symptoms of PSTVd natural infection in tomato plants expressing hairpin RNA derived from PSTVd, which has also been shown when artificial miRNA (amiRNA) corresponding to the virulence-modulating region of PSTVd had been used (Eamens et al. 2014; Wang et al. 2004). The significance of symptom induction by viroid sRNAs was also highlighted in Hop stunt viroid (HSVd)-infected tobacco plants deficient in RDR6 activity (Gómez et al., 2008). It was subsequently demonstrated that Peach latent mosaic viroid, (PLMVd) targets a host mRNA that encodes heat shock protein 90 (HSP90), leading to symptoms and suppression of host defense (Navarro et al., 2012). Viroid sRNAs from the virulence-modulating region of PSTVd target multiple host callose synthase genes, which are involved in callose deposition during pathogen infection (Adkar-Purushothama et al., 2015). A comparative genomics study for sRNAs from 29 pospiviroids identified a WD40-repeat gene of tomato as a target gene. In silico analysis of PSTVd sRNAs in tomatoes revealed three viroid sRNAs that target different transmembrane receptors involved in host defense. Furthermore, transient expression of the three viroid siRNAs induced phenotypes similar to viroid disease symptoms (Adkar-Purushothama and Perreault, 2018).
At the same time, viroids are capable of enhancing their replication by reprogramming the epigenetic RNA silencing of the host. The RNA of PSTVd has been found to physically interact with VIROID-BINDING PROTEIN 1 (VIRP1) in tomato, which contains a bromodomain, involved in epigenetic regulation (Martínez de Alba et al., 2002). HSVd also interacts physically with HISTONE DEACETYLASE 6 (HDA6), suggesting the promotion of transcriptional changes that enhance viroid replication (Castellano et al., 2016). Furthermore, alterations in phytohormone signaling and metabolic pathways were found in hop plants infected with Citrus bark cracking viroid (CBCVd) (Mishra et al., 2018). In silico predicted viroid sRNAs from eight pospiviroids were mapped to tomato coding sequences and the analysis of potential target genes revealed an over representation of gene ontology terms such as cytoskeleton, kinases and membrane transporters that can affect the fitness of pospiviroids or influence symptom development (Thibaut and Claude, 2018).
Besides the effect of viroids on host genes, they can also affect flowering. This has been shown in chrysanthemum plants infected with chrysanthemum stunt viroid as well as in tomato plants infected with PSTVd, in which flowering responsive genes were downregulated (Adkar-Purushothama et al., 2018). In line with the above observation, infection with tomato planta macho viroid (TPMVd) and Mexican papita viroid (MPVd) led to the alteration of the expression of two genes involved in fruit and flower development, OVA6 and BIGPETAL1, although the involvement of viroid siRNAs in this modulation has not been demonstrated (Aviña-Padilla et al., 2018).
Concluding remarks
It took several years for the scientific community to understand and unravel the role of RNA silencing in the regulation of endogenous genes expression as well as in the response of plants to various stimuli, either abiotic or biotic. Almost independently, plant and animal scientists reached the conclusion that there is a specific mechanism, which is triggered by the presence of dsRNAs, and leads to the repression of the complementary sequence in the genome (Fire et al. 1998; Lee et al. 1993; Reinhart et al. 2002; Stark et al. 2003). These dsRNAs could have diverse origin, varying from endogenous sequences with extensive fold-back structures to viral or viroid sequences and TEs. Depending on the origin of the trigger sequence, a specific RNA silencing pathway is activated, either the endogenous or the exogenous RNA silencing pathway (Liu et al. 2017; Pumplin and Voinnet 2013; Song et al. 2019). Each of the above pathways is guided by a specific classes of sRNAs that finally lead to transcriptional and post-transcriptional changes in the plant genome.
Probably the most studied class of sRNAs are the miRNAs, which guide the endogenous gene silencing pathway, regarded as one of the core mechanisms for the regulation of plant development, from leaf patterning to phase transition and response to environmental cues (Song et al., 2019; Waititu et al., 2020). At the same time, the role of RNA silencing has been extensively studied in relation to its role as a defense mechanism against biotic stress caused by the presence viruses and viroids (Garcia-Ruiz 2019; Pumplin and Voinnet 2013). In addition to the role of endogenous RNA silencing in the control and manipulation of plant growth, also of particular interest may be the involvement and interaction of miRNAs and endogenous silencing, in response to biotic stress adaptation of plants.
Acknowledgements
This work was financed by Greece and the European Union (European Social Fund-ESF) through the Operational Program «Human Resources Development, Education and Lifelong Learning» in the context of the project “Reinforcement of Postdoctoral Researchers-2nd Cycle” (MIS-5033021), implemented by the State Scholarships Foundation (ΙΚΥ). This publication reflects only the authors’ view. The Agency is not responsible for any use that may be made of the information it contains. This work was also supported by the project “Development of national research network to «Grape», Division 1. (Development of national research network to «Οlive», to «Grape», to «Honey», and to «Livestock»)”, with the code 2018ΣΕ01300000 of the National Scale of the Public Investments Program of the General Secretariat of Research and Innovation. In addition, this work was implemented in the framework of the Operation "DEVELOPMENT OF AN INTEGRATED SYSTEM FOR DETECTION OF PLANT PATHOGENS IN THE FIELD" with code OPS (MIS) 5022230 and is funded by the Operational Program "Crete 2014-20’’under the context of NSRF 2014-2020, co-financed by Greece and the European Union (European Structural and Investment Funds).
Abbreviations
AGOs, argonautes; CBP20 and CBP80, CAP-BINDING PROTEINS; DCL1-4, DICER-LIKE 1-4; DRB4, DOUBLE-STRANDED RNA BINDING FACTOR4; dsRNA, double-stranded RNA; dsRBD, double-stranded RNA binding domain; hcRNA, heterochromatic RNA; HEN1, HUA ENHANCER 1; HSVd, Hop stunt viroid; HYL1, HYPONASTIC LEAVES 1; miRNA, microRNA; nat-siRNA, natural antisense transcript-derived siRNA; ncRNA, non-coding RNA; pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA; PTGS, post-transcriptional gene silencing; Pol II, RNA polymerase II; Pol IV, RNA polymerase IV; Pol V, RNA polymerase V; PSTVd, Potato spindle tuber viroid; RdDM, RNA-directed DNA methylation; RISC, RNA-induced silencing complex; phasiRNAs, phased siRNAs; RNAi, RNA interference; RNAse III, Ribonuclease III; RDR1-6, RNA-DEPENDENT RNA POLYMERASE 1-6; SE, SERRATE; sRNA, small RNA; ssDNA, single-stranded DNA; ssRNA, single-stranded RNA; siRNA, small interfering RNA; tasiRNAs, trans-acting siRNAs; TGS, transcriptional gene silencing; TEs, transposable elements; vd-siRNAs, viroid-derived siRNAs; vsiRNAs, virus-derived small interfering RNAs.References
Adkar-Purushothama C. R., Kasai A., Sugawara K., Yamamoto H., Yamazaki Y., He Y.H., Takada N., Goto H., Shindo S., Harada T., Sano T. (2015). RNAi mediated inhibition of viroid infection in transgenic plants expressing viroid-specific small RNAs derived from various functional domains. Scientific Reports 5: 17949.
Adkar-Purushothama C. R., Sano T., Perreault J.P. (2018). Viroid-derived small RNA induces early flowering in tomato plants by RNA silencing. Molecular Plant Pathology 19: 2446-2458.
Mishra A., Kumar A., Mishra D., Nath V., Jakše J., Kocábek T., Killi U., Morina F., Matoušek J. (2018). Genome-Wide Transcriptomic Analysis Reveals Insights into the Response to Citrus bark cracking viroid (CBCVd) in Hop (Humulus lupulus L.). Viruses 10: 570.
Ali M., Javaid A., Naqvi S. H., Batcho A., Kayani W. K., Lal A., Sajid I. A., Nwogwugwu J. O. (2020). Biotic stress triggered small RNA and RNAi defense response in plants. Molecular Biology Reports 47: 5511-5522.
Allen E., Xie Z., Gustafson A. M., Carrington J. C. (2005). microRNA-Directed Phasing during Trans-Acting siRNA Biogenesis in Plants. Cell 121: 207-221.
Aviña-Padilla K., Rivera-Bustamante R., Kovalskaya N., Hammond R. (2018). Pospiviroid Infection of Tomato Regulates the Expression of Genes Involved in Flower and Fruit Development. Viruses 10: 516.
Bajczyk M., Lange H., Bielewicz D., Szewc L., Bhat S. S., Dolata J., Kuhn L., Szweykowska-Kulinska Z., Gagliardi D., Jarmolowski A. (2020). SERRATE interacts with the nuclear exosome targeting (NEXT) complex to degrade primary miRNA precursors in Arabidopsis. Nucleic Acids Research 48: 6839-6854.
Barciszewska-Pacak M., Knop K., Jarmołowski A., Szweykowska-Kulińska Z. (2016). Arabidopsis thaliana microRNA162 level is posttranscriptionally regulated via splicing and polyadenylation site selection.. Acta Biochimica Polonica 63: 811-816.
Baulcombe D. (2004). RNA silencing in plants. Nature 431: 356-363.
Bernstein E., Caudy A. A., Hammond S. M., Hannon G. J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366.
Bhogireddy S., Mangrauthia S. K., Kumar R., Pandey A. K., Singh S., Jain A., Budak H., Varshney R. K., Kudapa H. (2021). Regulatory non-coding RNAs: a new frontier in regulation of plant biology. Functional & Integrative Genomics 21: 313-330.
Borges F., Martienssen R. A. (2015). The expanding world of small RNAs in plants. Nature Reviews Molecular Cell Biology 16: 727-741.
Castellano M., Pallás V., Gómez G. (2016). A pathogenic long noncoding RNA redesigns the epigenetic landscape of the infected cells by subverting host Histone Deacetylase 6 activity. New Phytologist 211: 1311-1322.
Chen W., Zhang X., Fan Y., Li B., Ryabov E., Shi N., Zhao M., Yu Z., Qin C., Zheng Q., Zhang P., Wang H., Jackson S., Cheng Q., Liu Y., Gallusci P., Hong Y. (2018). A Genetic Network for Systemic RNA Silencing in Plants. Plant Physiology 176: 2700-2719.
Cheng J., Niu Q., Zhang B., Chen K., Yang R., Zhu J.K., Zhang Y., Lang Z. (2018). Downregulation of RdDM during strawberry fruit ripening. Genome Biology 19: 1-14.
Ciechanowska K., Pokornowska M., Kurzyńska-Kokorniak A. (2021). Genetic Insight into the Domain Structure and Functions of Dicer-Type Ribonucleases. International Journal of Molecular Sciences 22: 616.
Cuerda-Gil D., Slotkin R. K. (2016). Non-canonical RNA-directed DNA methylation. Nature Plants 2: 16163.
Dadami E., Boutla A., Vrettos N., Tzortzakaki S., Karakasilioti I., Kalantidis K. (2013). DICER-LIKE 4 But Not DICER-LIKE 2 May Have a Positive Effect on Potato Spindle Tuber Viroid Accumulation in Nicotiana benthamiana. Molecular Plant 6: 232-234.
Dalakouras A., Dadami E., Wassenegger M. (2015). Engineering Viroid Resistance. Viruses 7: 634-646.
Dalakouras A., Wassenegger M., Dadami E., Ganopoulos I., Pappas M. L., Papadopoulou K. (2020). Genetically Modified Organism-Free RNA Interference: Exogenous Application of RNA Molecules in Plants. Plant Physiology 182: 38-50.
Deng P., Muhammad S., Cao M., Wu L. (2018). Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnology Journal 16: 965-975.
Ding S.W., Voinnet O. (2007). Antiviral Immunity Directed by Small RNAs. Cell 130: 413-426.
Dong Z., Han M.H., Fedoroff N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1 . Proceedings of the National Academy of Sciences 105: 9970-9975.
Eamens A. L., Smith N. A., Dennis E. S., Wassenegger M., Wang M.B. (2014). In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of Potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology 450-451: 266-277.
Erdmann R. M., Picard C. L. (2020). RNA-directed DNA Methylation. PLOS Genetics 16: e1009034.
Fei Q., Xia R., Meyers B. C. (2013). Phased, Secondary, Small Interfering RNAs in Posttranscriptional Regulatory Networks. The Plant Cell 25: 2400-2415.
Fire A., Xu S.Q., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806-811.
Gao R., Wan Z. Y., Wong S.M. (2013). Plant Growth Retardation and Conserved miRNAs Are Correlated to Hibiscus Chlorotic Ringspot Virus Infection. PLoS ONE 8: e85476.
Gao S., Wang J., Jiang N., Zhang S., Wang Y., Zhang J., Li N., Fang Y., Yang L., Chen S., Yan B., Huang T., Kuai B., Wang Y., Chang F., Ren G. (2020). Hyponastic Leaves 1 protects pri-miRNAs from nuclear exosome attack. Proceedings of the National Academy of Sciences 117: 17429-17437.
Gao Z., Nie J., Wang H. (2021). MicroRNA biogenesis in plant. Plant Growth Regulation 93: 1-12.
Garcia‐Ruiz H. (2019). Host factors against plant viruses. Molecular Plant Pathology 20: 1588-1601.
Gómez G., Martínez G., Pallás V. (2008). Viroid-Induced Symptoms in Nicotiana benthamiana Plants Are Dependent on RDR6 Activity . Plant Physiology 148: 414-423.
Hamilton A. J., Baulcombe D. C. (1999). A Species of Small Antisense RNA in Posttranscriptional Gene Silencing in Plants. Science 286: 950-952.
Huang J., Yang M., Zhang X. (2016). The function of small RNAs in plant biotic stress response. Journal of Integrative Plant Biology 58: 312-327.
Hung Y.H., Slotkin R. K. (2021). The initiation of RNA interference (RNAi) in plants. Current Opinion in Plant Biology 61: 102014.
Itaya A., Zhong X., Bundschuh R., Qi Y., Wang Y., Takeda R., Harris A. R., Molina C., Nelson R. S., Ding B. (2007). A Structured Viroid RNA Serves as a Substrate for Dicer-Like Cleavage To Produce Biologically Active Small RNAs but Is Resistant to RNA-Induced Silencing Complex-Mediated Degradation. Journal of Virology 81: 2980-2994.
Jin Y., Zhao J.H., Guo H.S. (2020). Recent advances in understanding plant antiviral RNAi and viral suppressors of RNAi. Current Opinion in Virology 46: 65-72.
Kalantidis K., Psaradakis S., Tabler M., Tsagris M. (2002). The Occurrence of CMV-Specific Short RNAs in Transgenic Tobacco Expressing Virus-Derived Double-Stranded RNA is Indicative of Resistance to the Virus. Molecular Plant-Microbe Interactions® 15: 826-833.
Kalantidis K., Schumacher H. T., Alexiadis T., Helm J. M., (2008). RNA silencing movement in plants. Biology of the Cell 100: 13-26.
Katsarou K., Mavrothalassiti E., Dermauw W., Van Leeuwen T., Kalantidis K. (2016). Combined Activity of DCL2 and DCL3 Is Crucial in the Defense against Potato Spindle Tuber Viroid. PLOS Pathogens 12: e1005936.
Katsarou K., Mitta E., Bardani E., Oulas A., Dadami E., Kalantidis K. (2018). DCL-suppressed Nicotiana benthamiana plants: valuable tools in research and biotechnology . Molecular Plant Pathology 20: 432-446.
Katsarou K., Rao A.L.N., Tsagris M., Kalantidis K. (2015). Infectious long non-coding RNAs. Biochimie 117: 37-47.
van der Krol A. R., Mur L. A., Beld M., Mol J. N., Stuitje A. R. (1990). Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression.. The Plant Cell 2: 291-299.
Kryovrysanaki N., Alexiadis A., Grigoriadou A. M., Katsarou K., Kalantidis K. (2019). SERRATE, a miRNA biogenesis factor, affects viroid infection in Nicotiana benthamiana and Nicotiana tabacum. Virology 528: 164-175.
Lee R. C., Feinbaum R. L., Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843-854.
Li C., Zhang B. (2016). MicroRNAs in Control of Plant Development. Journal of Cellular Physiology 231: 303-313.
Li E., Zhang Y. (2014). DNA Methylation in Mammals. Cold Spring Harbor Perspectives in Biology 6: a019133-a019133.
Li S., Castillo‐González C., Yu B., Zhang X. (2017). The functions of plant small RNA s in development and in stress responses . The Plant Journal 90: 654-670.
Liu S.R., Zhou J.J., Hu C.G., Wei C.L., Zhang J.Z. (2017). MicroRNA-Mediated Gene Silencing in Plant Defense and Viral Counter-Defense. Frontiers in Microbiology 8: 1801.
Liu Y., Teng C., Xia R., Meyers B. C. (2020). PhasiRNAs in Plants: Their Biogenesis, Genic Sources, and Roles in Stress Responses, Development, and Reproduction. The Plant Cell 32: 3059-3080.
Lobbes D., Rallapalli G., Schmidt D. D., Martin C., Clarke J. (2006). SERRATE: a new player on the plant microRNA scene. EMBO reports 7: 1052-1058.
Martínez de Alba A. E., Flores R., Hernández C. (2002). Two Chloroplastic Viroids Induce the Accumulation of Small RNAs Associated with Posttranscriptional Gene Silencing. Journal of Virology 76: 13094-13096.
Matzke M. A., Mosher R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature Reviews Genetics 15: 394-408.
Megraw M., Baev V., Rusinov V., Jensen S. T., Kalantidis K., Hatzigeorgiou A. G. (2006). MicroRNA promoter element discovery in Arabidopsis . RNA 12: 1612-1619.
Mermigka G., Verret F., Kalantidis K. (2014). RNA silencing movement in plants. Journal of Integrative Plant Biology 58: 328-342.
Minoia S., Carbonell A., Di Serio F., Gisel A., Carrington J. C., Navarro B., Flores R. (2014). Specific Argonautes Selectively Bind Small RNAs Derived from Potato Spindle Tuber Viroid and Attenuate Viroid Accumulation In Vivo . Journal of Virology 88: 11933-11945.
Napoli C., Lemieux C., Jorgensen R. (1990). Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans.. The Plant Cell 2: 279-289.
Navarro B., Gisel A., Rodio M.E., Delgado S., Flores R., Di Serio F. (2012). Viroids: How to infect a host and cause disease without encoding proteins. Biochimie 94: 1474-1480.
Niaz S. (2018). The AGO proteins: an overview. Biological Chemistry 399: 525-547.
Pallás V., Martínez G., Gómez G. (2012). The Interaction Between Plant Viroid-Induced Symptoms and RNA Silencing. In Antiviral Resistance in Plants. (Ed. Watson John M., Wang Ming-Bo) Humana Press, Totowa, NJ.
Pantaleo V. (2011). Plant RNA Silencing in Viral Defence. In RNA Infrastructure and Networks. (Ed. Collins Lesley J.) Springer New York, New York, NY.
Pertermann R., Tamilarasan S., Gursinsky T., Gambino G., Schuck J., Weinholdt C., Lilie H., Grosse I., Golbik R. P., Pantaleo V., Behrens S.-E., (2018). A Viral Suppressor Modulates the Plant Immune Response Early in Infection by Regulating MicroRNA Activity. mBio 9: e00419-18.
Pooggin M. (2013). How Can Plant DNA Viruses Evade siRNA-Directed DNA Methylation and Silencing?. International Journal of Molecular Sciences 14: 15233-15259.
Pooggin M. M., Ryabova L. A. (2018). Ribosome Shunting, Polycistronic Translation, and Evasion of Antiviral Defenses in Plant Pararetroviruses and Beyond. Frontiers in Microbiology 9: 644.
Pumplin N., Voinnet O. (2013). RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nature Reviews Microbiology 11: 745-760.
Raczynska K. D., Stepien A., Kierzkowski D., Kalak M., Bajczyk M., McNicol J., Simpson C. G., Szweykowska-Kulinska Z., Brown J. W. S., Jarmolowski A. (2014). The SERRATE protein is involved in alternative splicing in Arabidopsis thaliana. Nucleic Acids Research 42: 1224-1244.
Rao A.L.N., Kalantidis K. (2015). Virus-associated small satellite RNAs and viroids display similarities in their replication strategies. Virology 479-480: 627-636.
Ray S., Golden T., Ray A. (1996). Maternal Effects of theshort integumentMutation on Embryo Development inArabidopsis. Developmental Biology 180: 365-369.
Rédei G. P., Hirono Y., (1964). Linkage studies. Arabidopsis Information Service 1: 9-10.
Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P. (2002). MicroRNAs in plants. Genes & Development 16: 1616-1626.
Sanan-Mishra N., Abdul Kader Jailani A., Mandal B., Mukherjee S. K. (2021). Secondary siRNAs in Plants: Biosynthesis, Various Functions, and Applications in Virology. Frontiers in Plant Science 12: 610283.
Sankaranarayanan R., Palani S. N., Kumar A., Selvakumar A. S. P., Tennyson J. (2020). Prediction and experimental confirmation of banana bract mosaic virus encoding miRNAs and their targets. ExRNA 2: 5.
Schauer S. E., Jacobsen S. E., Meinke D. W., Ray A. (2002). DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends in Plant Science 7: 487-491.
Schwind N., Zwiebel M., Itaya A., Ding B., Wang M., Krczal G., Wassenegger M. (2009). RNAi-mediated resistance to Potato spindle tuber viroid in transgenic tomato expressing a viroid hairpin RNA construct . Molecular Plant Pathology 10: 459-469.
Shen C., Wei C., Li J.n, Zhang X., Zhong Q., Li Y., Bai B., Wu Y., (2020). Barley yellow dwarf virus-GAV-derived vsiRNAs are involved in the production of wheat leaf yellowing symptoms by targeting chlorophyll synthase. Virology Journal 17: 158.
Smith H. A., Swaney S. L., Parks T. D., Wernsman E. A., Dougherty W. G. (1994). Transgenic plant virus resistance mediated by untranslatable sense RNAs: expression, regulation, and fate of nonessential RNAs.. The Plant Cell 6: 1441-1453.
Smith N. A., Eamens A. L., Wang M.B. (2011). Viral Small Interfering RNAs Target Host Genes to Mediate Disease Symptoms in Plants. PLoS Pathogens 7: e1002022.
Song X., Li Y., Cao X., Qi Y. (2019). MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annual Review of Plant Biology 70: 489-525.
Speth C., Szabo E. X., Martinho C., Collani S., zur Oven-Krockhaus S., Richter S., Droste-Borel I., Macek B., Stierhof Y.D., Schmid M., Liu C., Laubinger S. (2018). Arabidopsis RNA processing factor SERRATE regulates the transcription of intronless genes. eLife 7: e37078.
Stark A., Brennecke J., Russell R. B., Cohen S. M. (2003). Identification of Drosophila MicroRNA Targets. PLoS Biology 1: e60.
Stepien A., Knop K., Dolata J., Taube M., Bajczyk M., Barciszewska‐Pacak M., Pacak A., Jarmolowski A., Szweykowska‐Kulinska Z. (2017). Posttranscriptional coordination of splicing and miRNA biogenesis in plants . WIREs RNA 8: e1403.
Sunkar Ramanjulu, Li Yong-Fang, Jagadeeswaran Guru, (2012). Functions of microRNAs in plant stress responses. Trends in Plant Science 17: 196-203.
Thibaut O., Claude B. (2018). Innate Immunity Activation and RNAi Interplay in Citrus Exocortis Viroid—Tomato Pathosystem. Viruses 10: 587.
Thiebaut F., Hemerly A. S., Ferreira P. C. G. (2019). A Role for Epigenetic Regulation in the Adaptation and Stress Responses of Non-model Plants. Frontiers in Plant Science 10: 246.
Tsushima D., Adkar-Purushothama C. R., Taneda A., Sano T. (2014). Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild symptom-inducing isolates of Potato spindle tuber viroid. Journal of General Plant Pathology 81: 49-62.
Viswanathan C., Anburaj J., Prabu G. (2014). Identification and validation of sugarcane streak mosaic virus-encoded microRNAs and their targets in sugarcane. Plant Cell Reports 33: 265-276.
Waititu J. K., Zhang C., Liu J., Wang H. (2020). Plant Non-Coding RNAs: Origin, Biogenesis, Mode of Action and Their Roles in Abiotic Stress. International Journal of Molecular Sciences 21: 8401.
Wang J., Mei J., Ren G. (2019). Plant microRNAs: Biogenesis, Homeostasis, and Degradation. Frontiers in Plant Science 10: 360.
Wang M.B., Bian X.Y., Wu L.M., Liu L.X., Smith N. A., Isenegger D., Wu R.M., Masuta C., Vance V. B., Watson J. M., Rezaian A., Dennis E. S., Waterhouse P. M. (2004). On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proceedings of the National Academy of Sciences 101: 3275-3280.
Wang Z., Ma Z., Castillo-González C., Sun D., Li Y., Yu B., Zhao B., Li P., Zhang X. (2018). SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557: 516-521.
Wassenegger M., Dalakouras A. (2021). Viroids as a Tool to Study RNA-Directed DNA Methylation in Plants. Cells 10: 1187.
Waterhouse P. M., Graham M. W., Wang M.B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proceedings of the National Academy of Sciences 95: 13959-13964.
Willmann M. R., Endres M. W., Cook R. T., Gregory B. D. (2011). The Functions of RNA-Dependent RNA Polymerases in Arabidopsis. The Arabidopsis Book 9: e0146.
Wu H., Li B., Iwakawa H., Pan Y., Tang X., Ling-hu Q., Liu Y., Sheng S., Feng L., Zhang H., Zhang X., Tang Z., Xia X., Zhai J., Guo H. (2020). Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 581: 89-93.
Xu D., Zhou G., (2017). Characteristics of siRNAs derived from Southern rice black-streaked dwarf virus in infected rice and their potential role in host gene regulation. Virology Journal 14: 27.
Yan K., Liu P., Wu C.A., Yang G.D., Xu R., Guo Q.H., Huang J.G., Zheng C.C. (2012). Stress-Induced Alternative Splicing Provides a Mechanism for the Regulation of MicroRNA Processing in Arabidopsis thaliana. Molecular Cell 48: 521-531.
Yan Q., Cui X., Lin S., Gan S., Xing H., Dou D. (2016). GmCYP82A3, a Soybean Cytochrome P450 Family Gene Involved in the Jasmonic Acid and Ethylene Signaling Pathway, Enhances Plant Resistance to Biotic and Abiotic Stresses. PLOS ONE 11: e0162253.
Yang J., Zhang T., Li J., Wu N., Wu G., Yang J., Chen X., He L., Chen J., (2020). Chinese wheat mosaic virus ‐derived vsiRNA‐20 can regulate virus infection in wheat through inhibition of vacuolar‐ (H + )‐PPase induced cell death. New Phytologist 226: 205-220.
Yang T., Wang Y., Teotia S., Zhang Z., Tang G. (2018). The Making of Leaves: How Small RNA Networks Modulate Leaf Development. Frontiers in Plant Science 9: 824.
Yang X., Dong W., Ren W., Zhao Q., Wu F., He Y. (2021). Cytoplasmic HYL1 modulates miRNA-mediated translational repression. The Plant Cell 33: 1980-1996.
Yang Y., Liu T., Shen D., Wang J., Ling X., Hu Z., Chen T., Hu J., Huang J., Yu W., Dou D., Wang M.-B., Zhang B., (2019). Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLOS Pathogens 15: e1007534.
Yin Z., Chrzanowska M., Michalak K., Zimnoch-Guzowska E., (2014). Alteration of host-encoded miRNAs in virus infected plants—experimentally verified. In Plant Virus–Host Interaction. (Ed. Gaur R. K., Hohn T., Sharma P., ) Academic Press.
Yu Y., Jia T., Chen X. (2018). The ‘how’ and ‘where’ of plant micro RNA s . New Phytologist 216: 1002-1017.
Zhang H., Xia R., Meyers B. C., Walbot V. (2015). Evolution, functions, and mysteries of plant ARGONAUTE proteins. Current Opinion in Plant Biology 27: 84-90.
Zhang H., Lang Z., Zhu J.K. (2018a). Dynamics and function of DNA methylation in plants. Nature Reviews Molecular Cell Biology 19: 489-506.
Zhang R., Jing Y., Zhang H., Niu Y., Liu C., Wang J., Zen K., Zhang C.Y., Li D. (2018b). Comprehensive Evolutionary Analysis of the Major RNA-Induced Silencing Complex Members. Scientific Reports 8: 14189.
Zhang X., Lai T., Zhang P., Zhang X., Yuan C., Jin Z., Li H., Yu Z., Qin C., Tör M., Ma P., Cheng Q., Hong Y. (2019). Mini review: Revisiting mobile RNA silencing in plants. Plant Science 278: 113-117.
Zheng L., Zhang C., Shi C., Yang Z., Wang Y., Zhou T., Sun F., Wang H., Zhao S., Qin Q., Qiao R., Ding Z., Wei C., Xie L., Wu J., Li Y., (2017). Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice. PLOS Pathogens 13: e1006662.