Int. J. Dev. Biol. 66: 43 - 49 (2022)
Special Issue: Developmental Biology in Greece
The development of MGE-derived cortical interneurons: An Lhx6 tale
Open Access | Review | Published: 26 October 2021
Abstract
The cerebral cortex contains two main neuronal cell populations: the excitatory pyramidal neurons and the inhibitory interneurons, which constitute 20-30% of all cortical neurons. Cortical interneurons are characterized by a remarkable morphological, molecular and functional diversity. A swathe of research activity over the last 20 years has sought to determine how cortical interneurons acquire their mature cellular and functional features, and has identified a number of transcription factors that function at different stages of interneuron development. Here, we review all current knowledge concerning the multiple functions of the “master regulator” - LIM-Homeodomain transcription factor Lhx6 - a gene expressed in the medial ganglionic eminence of the basal telencephalon that controls the development of somatostatin and parvalbumin expressing interneurons.
Keywords
Lhx6, medial ganglionic eminence, cortical interneurons, parvalbumin, somatostatin, brain development
Introduction
GABAergic interneurons comprise 20-30% of all neurons in the cortex and are essential for cortical circuit function. They have multiple roles, from maintaining excitation/inhibition balance and synchronizing brain activity, to refining cortical processing in unique and multiple ways. Their functional diversity is enabled through their remarkable heterogeneity at the molecular, morphological and electrophysiological level. This diversity begins during embryonic development.
In mammals, most cortical and hippocampal interneurons are born in three distinct regions of the basal telencephalon: the medial ganglionic eminence (MGE), the caudal ganglionic eminence (CGE) and the preoptic area (POA) (Wonders and Anderson 2006; Kessaris et al. 2014; Lim et al. 2018). Each of these regions gives rise to distinct cohorts of interneurons of the cortex and hippocampus. The MGE is the largest source of interneurons for the cortex, generating around 60% of the total population. This includes two major classes: a) interneurons that are characterized by the expression of the calcium binding protein parvalbumin (PV), which exhibit fast-spiking firing properties, including basket, Chandelier and translaminar interneurons; and b) interneurons that are characterized by the expression of the neuropeptide somatostatin (SST) and the preferable dendritic targeting of their synapses. SST-expressing interneurons can be further subdivided into Martinotti, non-Martinotti and long-range GABAergic projection neurons.
Tremendous efforts have been made in the last 20 years to identify molecular mechanisms controlling interneuron development, after Anderson and colleagues discovered the origin of cortical interneurons in the embryonic subpallium (Anderson et al., 1997). Although our knowledge is still limited, details about the genetic cascades of transcription factors regulating MGE derived interneuron specification, migration and differentiation/maturation have begun to emerge. At the core of these gene networks is the LIM-homeodomain transcription factor Lhx6. In this review, we describe all current knowledge of the multiple functions of LHX6 at different stages of cortical interneuron development.
The LHX6/LHX8 group of LIM/homeodomain proteins
LIM/homeodomain genes encode transcription factors that are characterized by the association of two N-terminal LIM domains with a C-terminal homeodomain. The LIM motif encodes metal-binding cysteine and histidine-rich domains (Michelsen et al. 1994; Pérez-Alvarano et al. 1994, 1996; Konrat et al. 1997), and was first identified in the protein product of three genes: the nematode lin-11 (Freyd et al., 1990); mec-3 (Way and Chalfie, 1988); and the rat Islet-1 (Karlsson et al., 1990) genes. It has been suggested that the LIM domain functions as a protein-protein interaction motif (Schmeichel and Beckerle 1994; Arber and Caroni 1996), which regulates the binding specificity of the homeodomain to the DNA (Dawid et al., 1998). Several members of this family have been implicated in regulating specific aspects of patterning, neuronal identity, differentiation, and axon pathfinding in diverse tissues in both invertebrate and vertebrate organisms (Hobert and Westphal 2000).
LIM/homeodomain proteins can be subdivided into six classes, based on the sequence conservation of their homeodomain (Hobert and Westphal 2000). Lhx6 and Lhx8 (also termed L3 or Lhx7) (Grigoriou et al., 1998) belong to the most divergent class of LIM/homeodomain encoding genes (Hobert and Westphal 2000), which also includes the Drosophila arrowhead (Curtis et al., 1995) and the C.elegans lim-4 (Sagasti et al., 1999). Lhx6 and Lhx8 were first identified in a genetic screen performed to discover novel members of the LIM homeodomain family of transcription factors involved in the development of the nervous system (Grigoriou et al., 1998). During development, both genes are expressed in a highly overlapping pattern in the first branchial arch and the forebrain (Grigoriou et al., 1998).
Lhx6 and Lhx8 mRNAs are first detected in the oral ectomesenchyme of the maxillary and mandibular processes of the first branchial arch and the palatal shelves, at embryonic day (E)9.5. Their expression is progressively restricted to the mesenchyme of individual teeth, and is finally downregulated by postnatal day (P) 2 (Grigoriou et al. 1998; Zhao et al. 1999; Denaxa et al. 2009). Genetic loss of function studies using embryos deficient for both Lhx6 and Lhx7 highlighted their role in tooth development and patterning, as well as for normal cranial skeleton and palatal formation (Denaxa et al., 2009).
In the forebrain, expression of Lhx6 and Lhx8 is first detected around E11.5 in the MGE (Grigoriou et al., 1998). However, their expression is progressively restricted into distinct neuronal populations. In the adult mouse brain, Lhx6 is expressed in interneurons of the striatum and the cortex (see next section), whereas Lhx8 expression is maintained in cholinergic (ChAT) projection neurons of the basal forebrain, and ChAT+ interneurons of the striatum (Asbreuk et al. 2002; Zhao et al. 2003; Fragkouli et al. 2005). Analysis of several different Lhx8-deficient mouse lines revealed a requirement for LHX8 for the specification of these two neuronal types (Zhao et al. 2003; Mori et al. 2004, Fragkouli et al. 2005). Moreover, a number of studies established that LHX8 forms a hexamer with ISLET1, another LIM-homeodomain transcription factor, and together they determine the cholinergic neuronal fate in the vertebrate basal telencephalon (Fragkouli et al. 2009, Lopes et al. 2012; Cho et al. 2014).
Finally, Lhx6 and Lhx8 are also expressed in certain domains of the developing hypothalamus and distinct hypothalamic nuclei in the adult brain (Grigoriou et al. 1998; Blackshaw et al. 2010; Kim et al. 2021). LHX6 in particular has been shown to be implicated in an amygdalar-hypothalamic pathway controlling reproductive behaviours (Choi et al., 2005), and more recently in promoting sleep by controlling the function of a specific subpopulation of GABA-expressing neurons of the ventral zona incerta, in the hypothalamus (Liu et al., 2017).
Induction and Regulation of Lhx6 expression in the MGE
During embryogenesis, Lhx6 expression begins around E11.5 in the subventricular (SVZ) and mantle zone (MZ) of the MGE, while it is largely excluded from any other adjacent basal forebrain structures (Grigoriou et al., 1998). Lhx6 expression is directly activated by Nkx2.1 (Du et al. 2008; Sandeberg et al. 2016), a homeobox transcription factor that plays a fundamental role in the specification of the MGE neuroepithelium (Sussel et al., 1999). Nkx2.1 mutants show severe patterning defects in the basal forebrain and a dramatic loss of Lhx6, as well as Lhx8 and Shh expression, the latter being a known ventralising morphogen (Sussel et al., 1999). Interestingly, at later developmental stages, the network dynamics among these four genes change. Flandin et al., proposed a model in which LHX6 together with LHX8 promote the activation of the Shh gene in the same cells. SHH secretion from Lhx6/Lhx8-positive cells feeds forward onto the overlying ventricular zone (VZ) of the MGE, further promoting its identity, via the upregulation of Nkx2.1 and Nkx6.2 transcription factors (Flandin et al., 2011).
More recently, the CTCF (CCCTC-binding factor) genome organizing protein has been shown to regulate the expression of Lhx6 in the MGE. CTCF is not important for Lhx6 induction, since early Lhx6 expression is unaffected in Ctcf cKO embryos, but has a critical role in maintaining its optimal expression levels in immature GABAergic cells of the MGE. Several of these cells are known to co-express LHX8 (Fragkouli et al. 2009; Lopes et al. 2012). In Ctcf mutants, Lhx6 downregulation is complemented by an up-regulation of Lhx8 expression in the MGE, which most probably results in the change of fate of Lhx6+ GABAergic cells destined for the cortex, to Lhx8+ GABAergic projection neurons that remain in the basal forebrain (Elbert et al., 2019). Several CTCF binding sites have been identified in both Lhx6 and Lhx8 loci (Elbert et al., 2019). Thus, CTCF might regulate the choice between these two cell fates by gating a cross inhibitory regulation between Lhx6 and Lhx7.
The zinc-finger transcription factor Sp9 has been also been identified as a regulator of Lhx6 expression in the MGE. Chromatin immunoprecipitation sequencing (ChIP-Seq) experiments have identified several Sp9 peaks in the promoter and enhancer regions of Lhx6, while binding of Sp9 in one of these regulatory sequences activates transcription in an in vitro cell system (Liu et al., 2019). However, the complex phenotype of Sp9 cKO mice suggests that the modest decrease of Lhx6 expression that is observed in the MGE of the mutant mice might be partially responsible for the multiple defects of Sp9- MGE-derived cortical interneurons.
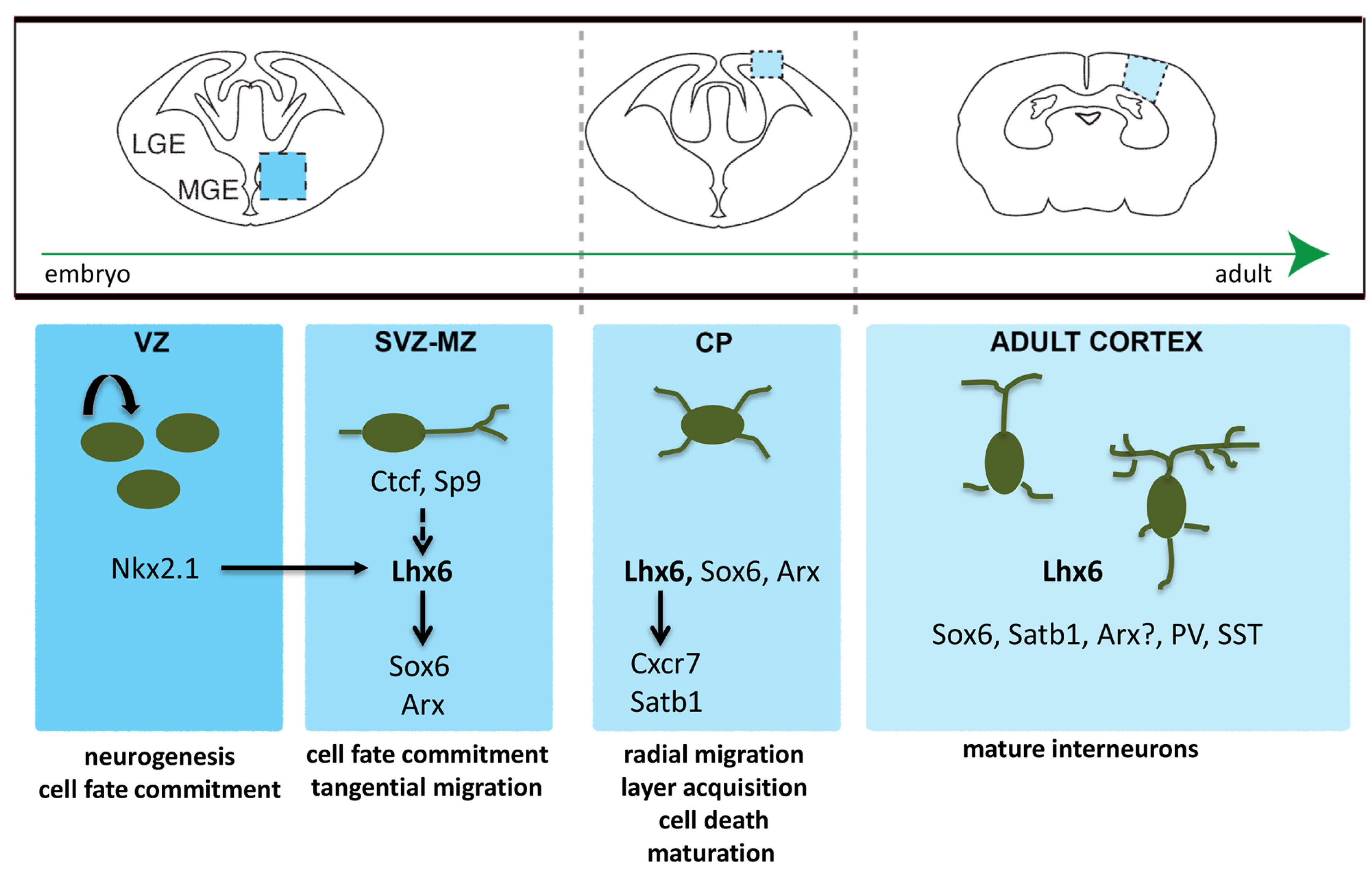
Nkx2.1 is only briefly expressed in the MGE lineage, and although it is required for the induction of Lhx6, its function is dispensable for the maintenance of Lhx6 expression (Sandeberg et al., 2016). On the other hand, Lhx6 expression is maintained in tangentially migrating interneurons (Lavdas et al. 1999; Liodis et al. 2007, Zhao et al. 2008), as well as in differentiated interneurons of the postnatal and adult brain. LHX6 is exclusively expressed in nearly 100% of MGE-derived PV-and SST-expressing interneurons within the adult cortex and hippocampus (Fogarty et al. 2007; Liodis et al. 2007; Zhao et al. 2008) (Fig. 1).
Fig. 1. A gene regulatory network for the development of MGE-derived PV- and SST-expressing cortical interneurons.
Nkx2.1 is required for the initiation of Lhx6 expression. Sp9 and Ctcf contributes to the maintenance of Lhx6 expression independently of Nkx2.1. Lhx6 promotes the expression of multiple genes (Sox6, Arx, Cxcr7, Satb1) that are subsequently required for the migration and maturation of cortical interneurons.
The role of Lhx6 in cortical interneuron development
Following description of the unique expression pattern of Lhx6 in the embryonic and adult telencephalon, several groups have begun to investigate its role in cortical interneuron development. The first loss of function data for Lhx6 resulted from small interfering RNA (siRNA) studies. Focal electroporation of an Lhx6 siRNA in the MGE of mouse embryonic brain slices inhibited the migration of GABAergic interneurons into the cortex (Alifragis et al., 2004).
Generation and analysis of the first Lhx6 mutant animals confirmed the above in vitro results, and further revealed that LHX6 controls not only the tangential but also the radial migration of immature interneurons into the cortex, as well as their differentiation into PV- and SST-expressing interneurons. More precisely, Lhx6-/- animals presented a delay in the front of migration during embryonic stages, but examination of their cortices at P15 revealed a similar number of GABA-expressing interneurons compared with controls, although their distribution into the various cortical layers was affected. In addition, Lhx6-null mutant mice displayed a severe reduction in both SST+ and PV+ interneuron numbers within the cortex (Liodis et al., 2007).
Zhao et al., using a different Lhx6 loss of function allele (Lhx6PLAP/PLAP), confirmed the above results and further identified additional molecular defects in Lhx6PLAP/PLAP interneurons, including a reduction in the Cxcr4 and Arx expression levels (Zhao et al., 2008; see also subsequent sections).
More recently, generation of a novel conditional allele of Lhx6 (Lhx6fl) in the mouse, allowed for MGE- specific Lhx6 ablation and simultaneous fate mapping of the MGE lineage. Notably, analysis of the above mice revealed that the number of MGE-derived interneurons was dramatically reduced in mutant mice compared with controls, suggesting that in addition to its previously described roles, Lhx6 is also required for the survival of MGE-derived interneurons. Indeed, further experiments revealed that Lhx6-deficient MGE-derived interneurons displayed a marked increase in apoptosis, even before the expected developmental cell death program begins (Denaxa et al., 2018a). These results, which initially seemed to contradict the original observations showing no significant loss of interneurons in the cortex of either Lhx6 null mutant mice described above (Liodis et al. 2007; Zhao et al. 2008), prompted the authors to investigate the lineage-identity of GABA-expressing interneurons that are found in Lhx6 mutant cortices. Fate mapping experiments across different developmental stages showed an increased representation of CGE-derived interneurons in the cortex of Lhx6 null mice, which was evident from early postnatal stages and was due to an increase in the fraction of CGE-derived interneurons that survive (Denaxa et al., 2018a). It is worth mentioning that this work was the first to present evidence of a non-cell autonomous mechanism controlling the survival of cortical interneurons (for review see Denaxa et al. 2018b; Wong and Marin 2019).
Downstream effectors of LHX6 function in cortical interneuron development
With the role of LHX6 in cortical interneuron development well established, several studies have been set up to unravel molecular cascades under LHX6 function (Denaxa et al. 2012; Sandberg et al. 2016). Here we describe the most prominent LHX6 effectors (Fig. 1).
The first gene described to act genetically downstream of LHX6 was the Sex Determining Region Y (SRY)-Box transcription factor SΟΧ6. Both histological analysis and genome-wide gene expression profiling revealed a dramatic decrease in Sox6 expression levels in the cortex of Lhx6 null compared with control mice (Batista-Brito et al. 2009; Denaxa et al. 2012). Similarly to Lhx6, Sox6 is expressed only in MGE-derived interneurons, and Sox6-deficient mice exhibit defects in the differentiation and function of PV+ and SST+ cortical interneurons (Azim et al. 2009; Batista-Brito et al. 2009). Notably, the number of PV-expressing interneurons was more severely affected than that of the SST positive interneuron subset, suggesting that Sox6 might play a more prominent role in the differentiation of PV+ cortical interneurons (Batista-Brito et al., 2009). Nevertheless, overexpression of Sox6 in Lhx6 mutant MGE cells transplanted into the cortex of control mice rescued neither PV or SST expression, while Sox6 overexpression into control MGE cells increased the percentage of SST- vs PV-expressing transplanted cortical interneurons (Vogt et al., 2014).
The Aristaless-related homeobox transcription factor (Arx) is another downstream effector of LHX6. Arx expression begins as early as E10.5 in the developing subpallium, and in contrast to Lhx6, its expression is not restricted only in the MGE, but is rather evident in all GE subdomains, as well as the POA (Cobos et al., 2005). As a result, Arx null mice are characterized by abnormal basal ganglia morphogenesis, and exhibit aberrant migration of GABA-expressing cells towards the cerebral cortex (Colombo et al., 2007). Histological and genome-wide gene expression experiments in Lhx6 null compared with control mice were the first to suggest that Lhx6 might be genetically upstream of Arx (Zhao et al. 2008; Denaxa et al. 2012). Subsequently, chromatin immunoprecipitation experiments clearly showed that LHX6 binds to enhancer elements near the Arx gene locus, while overexpression of ARX in Lhx6-deficient interneuron progenitors transplanted into control cortices rescued the expression of SST and PV, but failed to rescue their laminar distribution (Vogt et al., 2014).
As mentioned above, Lhx6 null mice are characterized by both a delay in their migration towards the cortex and an aberrant laminar distribution of GABA-expressing neurons. More specifically, MGE-derived cortical interneurons in Lhx6 mutants are lost in the middle cortical layers (IV) and are accumulated in superficial and deeper layers (I and V/VI) of the cortex (Liodis et al. 2007; Zhao et al. 2008; Denaxa et al. 2018). Previous studies have shown that these migratory events are mediated by chemokine signalling. CXCL12 (also known as stromal-derived factor 1, SDF-1) is a chemoattractant for cortical interneurons; its prenatal expression in the meninges and the intermediate and sub-ventricular zones (IZ/SVZ) of the developing cortex attracts immature cortical interneurons, expressing the receptors CXCR4 and CXCR7 and promoting their migration along the appropriate routes (Stumm et al. 2003; Lopez-Bendito et al. 2008; Sanchez-Alcanir et al. 2011; Wang et al. 2011). CXCR7 expression is greatly reduced in the MGE and migrating MGE-derived cells in Lhx6 mutants (Zhao et al., 2008). In addition, LHX6 directly binds to a Cxcr7 enhancer, and together with its cofactor Ldb1, modulates its activity in vitro. Finally, overexpression of CXCR7 partially rescued the laminar distribution of Lhx6-deficient MGE-derived cortical interneuron transplants (Vogt et al., 2014).
The special AT-rich sequence binding protein Satb1 has been found to be downregulated in a genome-wide gene expression profiling study on the dorsal forebrain of E15.5 Lhx6 mutants versus control littermates (Denaxa et al., 2012). Expression analysis in the adult cortex revealed that Satb1 expression is restricted in the MGE-derived lineage of cortical interneurons. In addition, analysis of two independently generated Satb1 mutant lines demonstrated a dramatic decrease in the number of SST- and to a lesser extent of PV-expressing cortical interneurons, in part due to cell death (Close et al. 2012; Denaxa et al. 2012). Furthermore, SATB1 overexpression in Lhx6 mutant MGE progenitors, which have been cultured on cortical feeder cells from control mice, partially rescued SST expression, while overexpression of a Satb1-specific ShRNA (Small Hairpin RNA) construct into control MGE cells using the same experimental set-up resulted in a dramatic reduction of SST-expressing interneurons. Although these results clearly demonstrated that Satb1 is genetically downstream of Lhx6, there were two pieces of evidence that clearly set Satb1 apart from any of the above-mentioned LHX6 effectors. First of all, both MGE progenitors, as well as tangentially migrating immature interneurons, lacked SATB1 expression, which initiated only in interneurons that have been already located in the cortical plate (CP), during late embryonic stages (Close et al. 2012; Denaxa et al. 2012). Secondly, overexpression of LHX6 in MGE precursors failed to up-regulate SATB1 expression. These observations, in combination with the demonstration that neuronal excitability is sufficient to upregulate SATB1 in control but not Lhx6-deficient cultured interneurons, suggested that LHX6 might control later stages of interneuron development by initiating a gene expression profile that allows these cells to respond to emerging environmental cues of the developing brain (Denaxa et al., 2012).
LHX6 and neurodevelopmental disorders
Loss of or aberrant cortical interneuron function has been associated with a number of neurodevelopmental disorders (Marin et al., 2012) (Table 1). It is not surprising therefore that Lhx6 null mutant animals display defects in the inhibitory circuits of the cortex and hippocampus. More specifically, Lhx6 null animals evidence a severe reduction in inhibitory synaptic currents, while hypomorphic Lhx6 mutants, which are characterized by aberrant defects in the number and function of SST- but not PV-positive interneurons, manifest spontaneous seizures from early postnatal stages (Neves et al., 2013).
Table 1
Transcription factors of the Lhx6 network and reported function in cortical interneuron development
| Transcription Factors | Mouse phenotype relevant to cortical interneuron development | Association with human psychiatric/disorders | References* |
| ARX | Aberrant migration and differentiation of cortical interneurons |
X-linked Mental Retardation with epilepsy and spasticity X-linked Lissencephaly with ambiguous genitalia X-linked Infantile Spasms syndrome Partington syndrome Proud syndrome West Syndrome |
Marsh and Golden, 2012 |
| CTCF |
Cell fate Migration Laminar Distribution |
Mental Retardation-21 | |
| LHX6 |
Cell fate Migration Laminar Distribution Survival Differentiation |
Schizophrenia Turret Syndrome |
|
| NKX2.1 |
Neuroepithelial patterning Cell fate Migration |
||
| SATB1 | Maturation | Autism | |
| SOX6 |
Migration Laminar Distribution Differentiation |
||
| SP9 |
Cell fate Migration |
*Literature providing insight into the association of the described transcription factors with psychiatric/neurological disorders
Similarly to rodents, the major source for cortical interneurons in humans and other primates is the basal telencephalon, which can be subdivided into analogous progenitor domains (Hansen et al. 2013; Ma et al. 2013). As in rodents, the human MGE is characterized by the expression of NKX2.1 and LHX6 (Hansen et al. 2013; Ma et al. 2013). In addition, LHX6 is expressed only in SST- and PV positive interneurons, as has been shown by histological analysis of human brain samples, as well as recent genome profiling experiments of human cortical neurons at the single cell level (Krienen et al., 2020). Although Lhx6 deficient mice develop seizures, LHX6 has not yet been associated with epilepsy in humans. Nevertheless, LHX6 is considered a biomarker for schizophrenia (SCZ), since numerous studies reported a reduction in LHX6 mRNA levels in the PFC of a subset of SCZ subjects (Volk et al. 2012, 2014, 2016; Bowen et al. 2019). Moreover, recent in silico studies have also identified a positive association between SNPs located in the 3’UTR region of LHX6 and Tourette Syndrome, a neurodevelopmental disorder with a very complex aetiology and pathophysiology (Pagliaroli et al., 2020).
Concluding Remarks
The emergence of novel sequencing methods that allow the profiling of transcriptome and chromatin states from thousands of individual neurons has revolutionized brain development. Several large-scale molecular profiling studies have already revealed a greater molecular diversity of interneurons than previously described (Huang and Paul, 2019). So far, these studies establish that Lhx6 and Nkx2.1 are foundational core transcripts for all interneuron identities arising from the MGE (Nowakowski et al. 2017; Huang and Paul, 2019). This coincides with work showing that LHX6 and NKX2.1 bind to common regulatory elements (REs) in the MGE that primarily mediate transcriptional activation (Sandberg et al., 2016). Thus, LHX6 together with NKX2.1 promote a transcriptionally permissive chromatin state and initiate a transcriptional program that controls fundamental molecular and cellular properties, which most probably are common to all MGE-derived interneuron subtypes. Nevertheless, LHX6 might also act together with different transcription factors to promote distinct MGE interneuron fates. For example, it has been suggested that LHX6 together with DLX1 promote the SST fate, whereas the combined function of LHX6 and DLX5/6 promotes the PV identity (Wang et al., 2010). Additionally, interesting findings in hypomorphic Lhx6 mutants, where only SST- but not PV-expressing interneurons are affected (Neves et al., 2013), suggest that differences in the dose requirements for LHX6 expression, therefore the differential binding of LHX6 to yet to be identified transcriptional cofactors, might also be implicated in MGE-derived interneuron cell fate decisions. Finally, Lhx6 expression is maintained in cortical and hippocampal PV- and SST-expressing interneurons during postnatal stages and in adulthood, the functional implications of which remain unknown. Further exploration of the molecular mechanisms underlying LHX6 function in the generation of interneuron types, as well as its potential role in the temporal progression of interneuron maturation, will enhance our understanding of genetic programs controlling cortical interneuron fate and function.
Acknowledgements
Financial support to OC, VA and MD, was provided by the Hellenic Foundation for Research and Innovation (ID 2564) and a Stavros Niarchos Foundation grant to B.S.R.C. “Alexander Fleming”, as part of the Foundation’s initiative to support the Greek research center ecosystem.
Abbreviations
CGE, Caudal Ganglionic Eminence ; ChAT, Choline Acetyltransferase ; CP, Cortical Plate ; GABA, γ-aminobuteric acid ; GE, Ganglionic Eminence ; IZ, Intermediate Zone ; LGE, Lateral Ganglionic Eminence ; MGE, Medial Ganglionic Eminence ; MZ, Mantle Zone ; POA, Preoptic Area ; PV, Parvalbumin ; SCZ, Schizophrenia ; siRNA, Small Interfering RNA ; SNP, Single Nucleotide Polymorphism ; SST, Somatostatin ; SVZ, SubVetricular Zone ; VZ, Ventricular Zone ;References
Alifragis P. (2004). Lhx6 Regulates the Migration of Cortical Interneurons from the Ventral Telencephalon But Does Not Specify their GABA Phenotype. Journal of Neuroscience 24: 5643-5648.
Anderson S. A., Eisenstat D. D., Shi L., Rubenstein J. L. R. (1997). Interneuron Migration from Basal Forebrain to Neocortex: Dependence on Dlx Genes . Science 278: 474-476.
Arber S., Caroni P. (1996). Specificity of single LIM motifs in targeting and LIM/LIM interactions in situ.. Genes & Development 10: 289-300.
Asbreuk CH, Van Schaick HS, Cox JJ, Kromkamp M, Smidt MP, Burbach JP., (2002). The homeobox genes Lhx7 and Gbx1 are expressed in the basal forebrain cholinergic system. Neuroscience 109: 287-298.
Azim E., Jabaudon D., Fame R. M., Macklis J. D. (2009). SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nature Neuroscience 12: 1238-1247.
Batista-Brito R., Rossignol E., Hjerling-Leffler J., Denaxa M., Wegner M., Lefebvre V., Pachnis V., Fishell G. (2009). The Cell-Intrinsic Requirement of Sox6 for Cortical Interneuron Development. Neuron 63: 466-481.
Blackshaw S., Scholpp S., Placzek M., Ingraham H., Simerly R., Shimogori T. (2010). Molecular Pathways Controlling Development of Thalamus and Hypothalamus: From Neural Specification to Circuit Formation. Journal of Neuroscience 30: 14925-14930.
Bowen E. F. W., Burgess J. L., Granger R., Kleinman J. E., Rhodes C. H. (2019). DLPFC transcriptome defines two molecular subtypes of schizophrenia. Translational Psychiatry 9: 147.
Cho H.H., Cargnin F., Kim Y., Lee B., Kwon R.J., Nam H., Shen R., Barnes A. P., Lee J. W., Lee S., Lee S.K. (2014). Isl1 Directly Controls a Cholinergic Neuronal Identity in the Developing Forebrain and Spinal Cord by Forming Cell Type-Specific Complexes. PLoS Genetics 10: e1004280.
Choi G. B., Dong H., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Swanson L. W., Anderson D. J. (2005). Lhx6 Delineates a Pathway Mediating Innate Reproductive Behaviors from the Amygdala to the Hypothalamus. Neuron 46: 647-660.
Close J., Xu H., De Marco Garcia N., Batista-Brito R., Rossignol E., Rudy B., Fishell G. (2012). Satb1 Is an Activity-Modulated Transcription Factor Required for the Terminal Differentiation and Connectivity of Medial Ganglionic Eminence-Derived Cortical Interneurons. Journal of Neuroscience 32: 17690-17705.
Cobos I., Long J. E., Thwin M. T., Rubenstein J. L. (2006). Cellular Patterns of Transcription Factor Expression in Developing Cortical Interneurons. Cerebral Cortex 16: i82-i88.
Colombo E., Collombat P., Colasante G., Bianchi M., Long J., Mansouri A., Rubenstein J. L. R., Broccoli V. (2007). Inactivation of Arx, the Murine Ortholog of the X-Linked Lissencephaly with Ambiguous Genitalia Gene, Leads to Severe Disorganization of the Ventral Telencephalon with Impaired Neuronal Migration and Differentiation. Journal of Neuroscience 27: 4786-4798.
Curtiss J., Heilig J.S. (1995). Establishment of Drosophila imaginal precursor cells is controlled by the Arrowhead gene. Development 121: 3819-3828.
Dawid I. B., (1998). LIM protein interactions: Drosophila enters the stage. Trends in genetics : TIG 14: 480-482.
den Hoed J., de Boer E., Voisin N., Dingemans A. J.M., Guex N., Wiel L., Nellaker C., Amudhavalli S. M., Banka S., Bena F. S., Ben-Zeev B., Bonagura V. R., Bruel A.L., Brunet T., Brunner H. G., Chew H. B., Chrast J., Cimbalistienė L., Coon H., Délot E. C., Démurger F., Denommé-Pichon A.S., Depienne C., Donnai D., Dyment D. A., Elpeleg O., Faivre L., Gilissen C., Granger L., Haber B., Hachiya Y., Abedi Y. H., Hanebeck J., Hehir-Kwa J. Y., Horist B., Itai T., Jackson A., Jewell R., Jones K. L., Joss S., Kashii H., Kato M., Kattentidt-Mouravieva A. A., Kok F., Kotzaeridou U., Krishnamurthy V., Kučinskas V., Kuechler A., Lavillaureix A., Liu P., Manwaring L., Matsumoto N., Mazel B., McWalter K., Meiner V., Mikati M. A., Miyatake S., Mizuguchi T., Moey L. H., Mohammed S., Mor-Shaked H., Mountford H., Newbury-Ecob R., Odent S., Orec L., Osmond M., Palculict T. B., Parker M., Petersen A. K., Pfundt R., Preikšaitienė E., Radtke K., Ranza E., Rosenfeld J. A., Santiago-Sim T., Schwager C., Sinnema M., Snijders Blok L., Spillmann R. C., Stegmann A. P.A., Thiffault I., Tran L., Vaknin-Dembinsky A., Vedovato-dos-Santos J. H., Schrier Vergano S. A., Vilain E., Vitobello A., Wagner M., Waheeb A., Willing M., Zuccarelli B., Kini U., Newbury D. F., Kleefstra T., Reymond A., Fisher S. E., Vissers L. E.L.M. (2021). Mutation-specific pathophysiological mechanisms define different neurodevelopmental disorders associated with SATB1 dysfunction. The American Journal of Human Genetics 108: 346-356.
Denaxa M., Kalaitzidou M., Garefalaki A., Achimastou A., Lasrado R., Maes T., Pachnis V. (2012). Maturation-Promoting Activity of SATB1 in MGE-Derived Cortical Interneurons. Cell Reports 2: 1351-1362.
Denaxa M., Neves G., Rabinowitz A., Kemlo S., Liodis P., Burrone J., Pachnis V. (2018a). Modulation of Apoptosis Controls Inhibitory Interneuron Number in the Cortex. Cell Reports 22: 1710-1721.
Denaxa M., Neves G., Burrone J., Pachnis V. (2018b). Homeostatic Regulation of Interneuron Apoptosis During Cortical Development. Journal of Experimental Neuroscience 12: 117906951878427.
Denaxa M., Sharpe P. T., Pachnis V. (2009). The LIM homeodomain transcription factors Lhx6 and Lhx7 are key regulators of mammalian dentition. Developmental Biology 333: 324-336.
Du T., Xu Q., Ocbina P. J., Anderson S. A. (2008). NKX2.1 specifies cortical interneuron fate by activating Lhx6 . Development 135: 1559-1567.
Elbert A., Vogt D., Watson A., Levy M., Jiang Y., Brûlé E., Rowland M. E., Rubenstein J., Bérubé N. G. (2019). CTCF Governs the Identity and Migration of MGE-Derived Cortical Interneurons. The Journal of Neuroscience 39: 177-192.
Flandin P., Zhao Y., Vogt D., Jeong J., Long J., Potter G., Westphal H., Rubenstein J. L.R. (2011). Lhx6 and Lhx8 Coordinately Induce Neuronal Expression of Shh that Controls the Generation of Interneuron Progenitors. Neuron 70: 939-950.
Fogarty M., Grist M., Gelman D., Marin O., Pachnis V., Kessaris N. (2007). Spatial Genetic Patterning of the Embryonic Neuroepithelium Generates GABAergic Interneuron Diversity in the Adult Cortex. Journal of Neuroscience 27: 10935-10946.
Fragkouli A., Hearn C., Errington M., Cooke S., Grigoriou M., Bliss T., Stylianopoulou F., Pachnis V. (2005). Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. European Journal of Neuroscience 21: 2923-2938.
Fragkouli A., van Wijk N. V., Lopes R., Kessaris N., Pachnis V. (2009). LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development 136: 3841-3851.
Freyd G., Kim S. K., Horvitz H. R. (1990). Novel cysteine-rich motif and homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-II. Nature 344: 876-879.
Gregor A., Oti M., Kouwenhoven E. N., Hoyer J., Sticht H., Ekici A. B., Kjaergaard S., Rauch A., Stunnenberg H. G., Uebe S., Vasileiou G., Reis A., Zhou H., Zweier C. (2013). De Novo Mutations in the Genome Organizer CTCF Cause Intellectual Disability. The American Journal of Human Genetics 93: 124-131.
Grigoriou M., Tucker A.S., Sharpe P.T., Pachnis V. (1998). Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development 125: 2063-2074.
Hansen D. V., Lui J. H., Flandin P., Yoshikawa K., Rubenstein J. L., Alvarez-Buylla A., Kriegstein A. R. (2013). Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nature Neuroscience 16: 1576-1587.
Hobert O., Westphal H., (2000). Functions of LIM-homeobox genes. Trends in genetics : TIG 16: 75-83.
Huang Z. J., Paul A. (2019). The diversity of GABAergic neurons and neural communication elements. Nature Reviews Neuroscience 20: 563-572.
Janowski M., Milewska M., Zare P., Pękowska A. (2021). Chromatin Alterations in Neurological Disorders and Strategies of (Epi)Genome Rescue. Pharmaceuticals 14: 765.
Karlsson O., Thor S., Norberg T., Ohlsson H., Edlund T. (1990). Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo-and a Cys–His domain. Nature 344: 879-882.
Kessaris N., Magno L., Rubin A. N., Oliveira M. G. (2014). Genetic programs controlling cortical interneuron fate. Current Opinion in Neurobiology 26: 79-87.
Kim D. W., Liu K., Wang Z. Q., Zhang Y. S., Bathini A., Brown M. P., Lin S. H., Washington P. W., Sun C., Lindtner S., Lee B., Wang H., Shimogori T., Rubenstein J. L. R., Blackshaw S. (2021). Gene regulatory networks controlling differentiation, survival, and diversification of hypothalamic Lhx6-expressing GABAergic neurons. Communications Biology 4: 95.
Konrat R., Weiskirchen R., Kräutler B., Bister K. (1997). Solution Structure of the Carboxyl-terminal LIM Domain from Quail Cysteine-rich Protein CRP2. Journal of Biological Chemistry 272: 12001-12007.
Krienen F. M., Goldman M., Zhang Q., C. H. del Rosario R., Florio M., Machold R., Saunders A., Levandowski K., Zaniewski H., Schuman B., Wu C., Lutservitz A., Mullally C. D., Reed N., Bien E., Bortolin L., Fernandez-Otero M., Lin J. D., Wysoker A., Nemesh J., Kulp D., Burns M., Tkachev V., Smith R., Walsh C. A., Dimidschstein J., Rudy B., S. Kean L., Berretta S., Fishell G., Feng G., McCarroll S. A. (2020). Innovations present in the primate interneuron repertoire. Nature 586: 262-269.
Lavdas A. A., Grigoriou M., Pachnis V., Parnavelas J. G. (1999). The Medial Ganglionic Eminence Gives Rise to a Population of Early Neurons in the Developing Cerebral Cortex. The Journal of Neuroscience 19: 7881-7888.
Lim L., Mi D., Llorca A., Marín O. (2018). Development and Functional Diversification of Cortical Interneurons. Neuron 100: 294-313.
Liodis P., Denaxa M., Grigoriou M., Akufo-Addo C., Yanagawa Y., Pachnis V. (2007). Lhx6 Activity Is Required for the Normal Migration and Specification of Cortical Interneuron Subtypes. Journal of Neuroscience 27: 3078-3089.
Liu K., Kim J., Kim D. W., Zhang Y. S., Bao H., Denaxa M., Lim S.A., Kim E., Liu C., Wickersham I. R., Pachnis V., Hattar S., Song J., Brown S. P., Blackshaw S. (2017). Lhx6-positive GABA-releasing neurons of the zona incerta promote sleep. Nature 548: 582-587.
Liu Z., Zhang Z., Lindtner S., Li Z., Xu Z., Wei S., Liang Q., Wen Y., Tao G., You Y., Chen B., Wang Y., Rubenstein J. L., Yang Z. (2019). Sp9 Regulates Medial Ganglionic Eminence-Derived Cortical Interneuron Development. Cerebral Cortex 29: 2653-2667.
Lopes R., Verhey van Wijk N., Neves G., Pachnis V. (2012). Transcription factor LIM homeobox 7 (Lhx7) maintains subtype identity of cholinergic interneurons in the mammalian striatum. Proceedings of the National Academy of Sciences 109: 3119-3124.
Lopez-Bendito G., Sanchez-Alcaniz J. A., Pla R., Borrell V., Pico E., Valdeolmillos M., Marin O. (2008). Chemokine Signaling Controls Intracortical Migration and Final Distribution of GABAergic Interneurons. Journal of Neuroscience 28: 1613-1624.
Ma T., Wang C., Wang L., Zhou X., Tian M., Zhang Q., Zhang Y., Li J., Liu Z., Cai Y., Liu F., You Y., Chen C., Campbell K., Song H., Ma L., Rubenstein J. L., Yang Z. (2013). Subcortical origins of human and monkey neocortical interneurons. Nature Neuroscience 16: 1588-1597.
Marín O., (2012). Interneuron dysfunction in psychiatric disorders. Nature Reviews Neuroscience 13: 107-120.
Marsh E. D., Golden J. A., (2012). Developing Models of Aristaless-related homeobox mutations. In Jasper's Basic Mechanisms of the Epilepsies. (Ed. Noebels J. L., Avoli M., Rogawski M. A., Olsen R. W., Delgado-Escueta A. V., ) National Center for Biotechnology Information, Bethesda.
Michelsen J.W., Sewell A.K., Louis H.A., Olsen J.I., Davis D.R., Winge D.R., Beckerle M.C. (1994). Mutational analysis of the metal sites in an LIM domain. Journal of Biological Chemistry 269: 11108-11113.
Mori T., Yuxing Z., Takaki H., Takeuchi M., Iseki K., Hagino S., Kitanaka J., Takemura M., Misawa H., Ikawa M., Okabe M., Wanaka A. (2004). The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. European Journal of Neuroscience 19: 3129-3141.
Neves G., Shah M. M., Liodis P., Achimastou A., Denaxa M., Roalfe G., Sesay A., Walker M. C., Pachnis V. (2013). The LIM Homeodomain Protein Lhx6 Regulates Maturation of Interneurons and Network Excitability in the Mammalian Cortex. Cerebral Cortex 23: 1811-1823.
Nowakowski T. J., Bhaduri A., Pollen A. A., Alvarado B., Mostajo-Radji M. A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S. J., Velmeshev D., Ounadjela J. R., Shuga J., Wang X., Lim D. A., West J. A., Leyrat A. A., Kent W. J., Kriegstein A. R. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358: 1318-1323.
Pagliaroli L., Vereczkei A., Padmanabhuni S. S., Tarnok Z., Farkas L., Nagy P., Rizzo R., Wolanczyk T., Szymanska U., Kapisyzi M., Basha E., Koumoula A., Androutsos C., Tsironi V., Karagiannidis I., Paschou P., Barta C. (2020). Association of Genetic Variation in the 3'UTR of LHX6, IMMP2L, and AADAC With Tourette Syndrome. Frontiers in Neurology 11: 803.
Parikshak N. N., Luo R., Zhang A., Won H., Lowe J. K., Chandran V., Horvath S., Geschwind D. H. (2013). Integrative Functional Genomic Analyses Implicate Specific Molecular Pathways and Circuits in Autism. Cell 155: 1008-1021.
Pérez-Alvarado G. C., Kosa J. L., Louis H. A., Beckerle M. C., Winge D. R., Summers M. F., (1996). Structure of the cysteine-rich intestinal protein, CRIP. Journal of molecular biology 257: 153-174.
Pérez-Alvarado G. C., Miles C., Michelsen J. W., Louis H. A., Winge D. R., Beckerle M. C., Summers M. F. (1994). Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nature Structural & Molecular Biology 1: 388-398.
Sagasti A., Hobert O., Troemel E. R., Ruvkun G., Bargmann C. I. (1999). Alternative olfactory neuron fates are specified by the LIM homeobox gene lim-4. Genes & Development 13: 1794-1806.
Sánchez-Alcañiz J. A., Haege S., Mueller W., Pla R., Mackay F., Schulz S., López-Bendito G., Stumm R., Marín O. (2011). Cxcr7 Controls Neuronal Migration by Regulating Chemokine Responsiveness. Neuron 69: 77-90.
Sandberg M., Flandin P., Silberberg S., Su-Feher L., Price J. D., Hu J. S., Kim C., Visel A., Nord A. S., Rubenstein J. L.R. (2016). Transcriptional Networks Controlled by NKX2-1 in the Development of Forebrain GABAergic Neurons. Neuron 91: 1260-1275.
Schmeichel K. (1994). The LIM domain is a modular protein-binding interface. Cell 79: 211-219.
Stumm R. K., Zhou C., Ara T., Lazarini F., Dubois-Dalcq M., Nagasawa T., Höllt V., Schulz S. (2003). CXCR4 Regulates Interneuron Migration in the Developing Neocortex. The Journal of Neuroscience 23: 5123-5130.
Sussel L., Marin O., Kimura S., Rubenstein J.L. (1999). Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development 126: 3359-3370.
Vogt D., Hunt R. F., Mandal S., Sandberg M., Silberberg S. N., Nagasawa T., Yang Z., Baraban S. C., Rubenstein J. L.R. (2014). Lhx6 Directly Regulates Arx and CXCR7 to Determine Cortical Interneuron Fate and Laminar Position. Neuron 82: 350-364.
Volk D. W., Edelson J. R., Lewis D. A. (2014). Cortical Inhibitory Neuron Disturbances in Schizophrenia: Role of the Ontogenetic Transcription Factor Lhx6. Schizophrenia Bulletin 40: 1053-1061.
Volk D. W., Matsubara T., Li S., Sengupta E. J., Georgiev D., Minabe Y., Sampson A., Hashimoto T., Lewis D. A. (2012). Deficits in Transcriptional Regulators of Cortical Parvalbumin Neurons in Schizophrenia. American Journal of Psychiatry 169: 1082-1091.
Volk D. W., Sampson A. R., Zhang Y., Edelson J. R., Lewis D. A. (2016). Cortical GABA markers identify a molecular subtype of psychotic and bipolar disorders. Psychological Medicine 46: 2501-2512.
Wang Y., Dye C. A., Sohal V., Long J. E., Estrada R. C., Roztocil T., Lufkin T., Deisseroth K., Baraban S. C., Rubenstein J. L. R., (2010). Dlx5 and Dlx6 Regulate the Development of Parvalbumin-Expressing Cortical Interneurons. Journal of Neuroscience 30: 5334-5345.
Wang Y., Li G., Stanco A., Long J. E., Crawford D., Potter G. B., Pleasure S. J., Behrens T., Rubenstein J. L.R. (2011). CXCR4 and CXCR7 Have Distinct Functions in Regulating Interneuron Migration. Neuron 69: 61-76.
Way J. C., Chalfie M. (1988). mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54: 5-16.
Wonders C. P., Anderson S. A. (2006). The origin and specification of cortical interneurons. Nature Reviews Neuroscience 7: 687-696.
Wong F. K., Marín O. (2019). Developmental Cell Death in the Cerebral Cortex. Annual Review of Cell and Developmental Biology 35: 523-542.
Zhao Y., Flandin P., Long J. E., Cuesta M. D., Westphal H., Rubenstein J. L.R. (2008). Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. The Journal of Comparative Neurology 510: 79-99.
Zhao Y., Guo Y.J., Tomac A. C., Taylor N. R., Grinberg A., Lee E. J., Huang S., Westphal H. (1999). Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene Lhx8. Proceedings of the National Academy of Sciences 96: 15002-15006.
Zhao Y., Marin O., Hermesz E., Powell A., Flames N., Palkovits M., Rubenstein J. L. R., Westphal H. (2003). The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proceedings of the National Academy of Sciences 100: 9005-9010.