Int. J. Dev. Biol. 68: 231 - 239 (2024)
Special Issue: Developmental Biology in Nordic Countries
Wnt target gene Ascl4 is dispensable for skin appendage development
Open Access | Original Article | Published: 13 May 2024
Abstract
The development of skin appendages, including hair follicles, teeth and mammary glands is initiated through the formation of the placode, a local thickening of the epithelium. The Wnt/β-catenin signaling cascade is an evolutionary conserved pathway with an essential role in placode morphogenesis, but its downstream targets and their exact functions remain ill defined. In this study, we identify Achaete-scute complex-like 4 (Ascl4) as a novel target of the Wnt/β-catenin pathway and demonstrate its expression pattern in the signaling centers of developing hair follicles and teeth. Ascl transcription factors belong to the superfamily of basic helix-loop-helix transcriptional regulators involved in cell fate determination in many tissues. However, their specific role in the developing skin remains largely unknown. We report that Ascl4 null mice have no overt phenotype. Absence of Ascl4 did not impair hair follicle morphogenesis or hair shaft formation suggesting that it is non-essential for hair follicle development. No tooth or mammary gland abnormalities were detected either. We suggest that other transcription factors may functionally compensate for the absence of Ascl4, but further research is warranted to assess this possibility.
Keywords
hair follicle, mammary gland, tooth, transcription factor, Wnt
Introduction
The development of ectodermal appendages, such as hair follicles, teeth and various glands, shares a set of well-defined stages, including induction, placode and bud stages (Mikkola and Millar, 2006). In the early stages of development, reciprocal and sequential tissue interactions between the ectodermal epithelium and the underlying mesenchyme result in the formation of an epithelial thickening, or placode, serving as a precursor for the mature ectodermal appendage (Biggs and Mikkola, 2014; Mikkola and Millar, 2006). Once placodes are established, the epithelium expands into the condensing mesenchyme to form a bud, followed by organ-specific epithelial morphogenesis. The specific details of the later morphogenetic and differentiation programs vary widely among different organs, reflecting the diverse forms and functions of fully formed ectodermal appendages (Biggs and Mikkola, 2014). The invaginating hair bud progresses via peg and bulbous peg stages, concomitant with gradual differentiation of the follicular cell types, to produce the mature hair follicle that undergoes cyclic regeneration throughout the lifetime (Sennett and Rendl, 2012). Tooth morphogenesis, on the other hand, proceeds through the cap and bell stages, prior to the onset of mineralization of the hard tissues and root formation (Balic and Thesleff, 2015). The mammary bud initially grows slowly, but when it reaches the adjacent fat pad precursor tissue (prospective adult stroma) it undergoes branching morphogenesis generating a small ductal tree by birth (Spina and Cowin, 2021).
Ectodermal appendage development is orchestrated by a limited number of conserved signaling cascades, encompassing both shared and unique pathways for each organ (Mikkola and Millar, 2006). Members of the Wnt, transforming growth factor β (Tgfβ), fibroblast growth factor (Fgf), hedgehog (Hh), and tumor necrosis factor (Tnf) families have been extensively studied during ectodermal organ formation using genetically engineered mouse models. However, the downstream events and target genes remain incompletely characterized (Biggs and Mikkola, 2014; Yu and Klein, 2020). The canonical Wnt pathway, mediated by β-catenin and Tcf/Lef1 family of transcription factors, is an evolutionary conserved pathway with essential roles in tissue morphogenesis and cell fate specification (Loh et al., 2016). In developing skin appendages, it plays a crucial role in the early inductive events. The suppression of canonical Wnt signaling in the epidermis, achieved either by overexpression of the secreted Wnt inhibitor Dkk1 or epithelial deletion of β-catenin, leads to absence of all signs of hair and mammary placode formation (Andl et al., 2002; Chu et al., 2004; Huelsken et al., 2001; Zhang et al., 2009), and tooth development halts at the placode stage (Liu et al., 2008). On the other hand, forced epithelial β-catenin activation in the epidermis leads to precocious induction of hair follicle development, committing the entire skin to hair follicle fate (Närhi et al., 2008; Zhang et al., 2008), and in the oral epithelium, results in the formation of supernumerary teeth (Järvinen et al., 2006; Liu et al., 2008).
The acquisition of specific morphogenetic programs not only requires essential signaling molecules but also distinct combinations of transcription factors that elicit cell fate changes during embryonic development (Iwafuchi-Doi and Zaret, 2016). Ascl4 belongs to the achaete-scute complex-like family of transcription factors (Ascl1-5), characterized by a conserved basic-helix-loop-helix DNA-binding domain (García-Bellido and De Celis, 2009). Various Ascl family members are involved in cell fate determination in different tissues such as Ascl1 in neural fate commitment (Guillemot and Hassan, 2017), and Ascl2 in intestinal stem cell and trophoblast cell lineage specification (Lefebvre, 2012; van der Flier et al., 2009). However, knowledge about Ascl4 is limited, although it is known to be expressed in the embryonic human skin (Jonsson et al., 2004).
Recently, we transcriptionally profiled mouse skin epithelial cells at embryonic day 14.5 (E14.5) when the first hair placodes have just formed. Our bulk RNA sequencing (RNA-seq) data, confirmed by unbiased single-cell RNA-seq (scRNA-seq) profiling, revealed that Ascl4 is highly enriched in the hair placode population compared to the interfollicular epidermis (Sulic et al., 2023), consistent with findings from other hair placode profiling studies (Sennett et al., 2015; Tomann et al., 2016). In this study, we investigated the role of Ascl4 in ectodermal appendage development, specifically focusing on hair follicles, teeth, and mammary gland. Our findings establish a connection between Ascl4 and Wnt signaling, showing that Ascl4 is a likely direct transcriptional target of the Wnt pathway. However, our results indicate that Ascl4 is dispensable for ectodermal organogenesis.
Results
Ascl4 is expressed in hair and tooth signaling centers
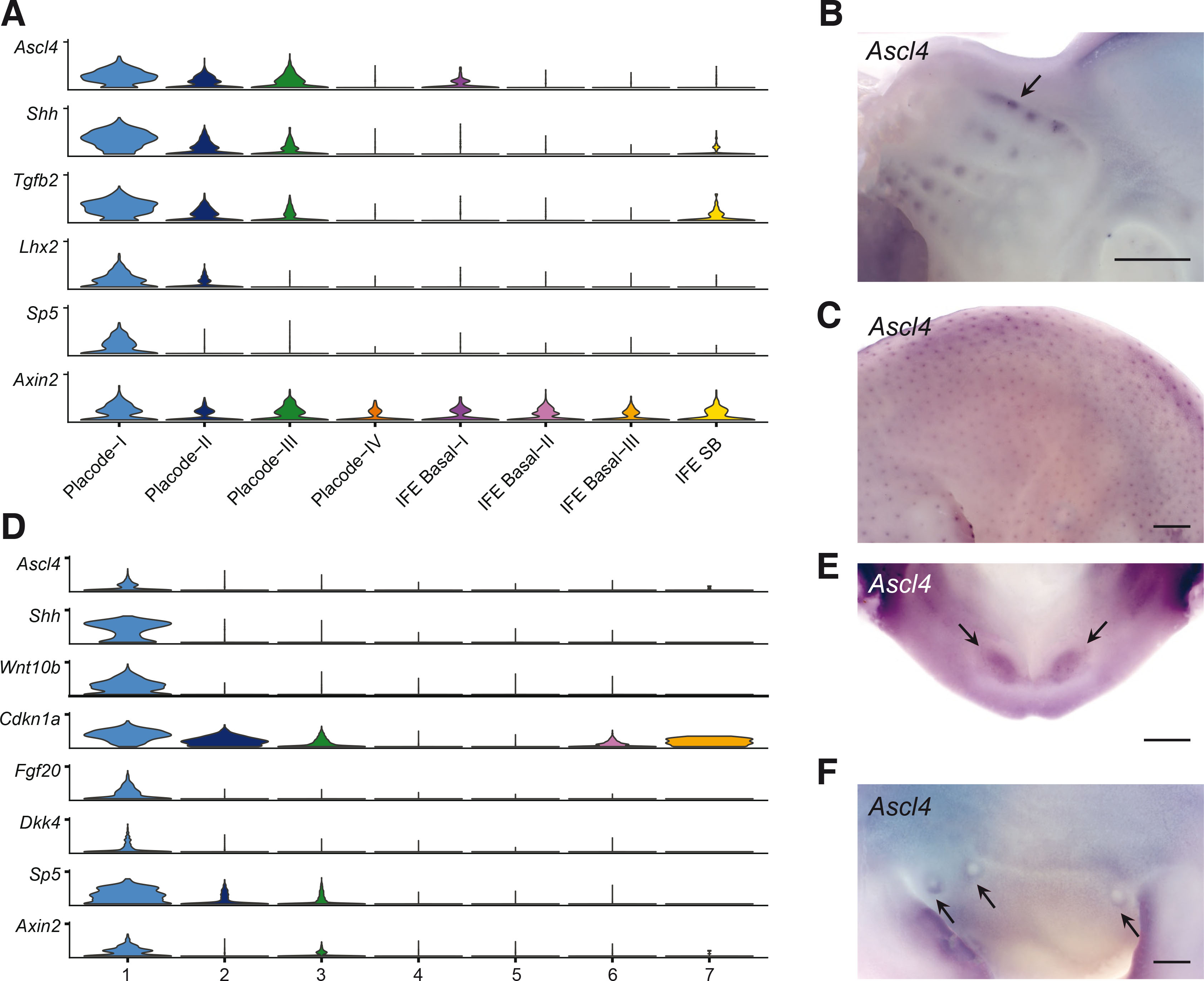
Our recent scRNA-seq profiling of hair placode-enriched cells identified four spatially distinct cell populations: one representing a central population (Placode-I), another one at the placode edge (Placode-IV), and two populations located in-between (Placode-II and Placode-III). Intriguingly, Ascl4 was predominantly expressed in the Placode-I population (Fig. 1A; Sulic et al., 2023). This cell population is defined by high expression of signaling molecules such as Tgfβ2, Bmp2, and Shh. Other markers include Tgfb2 and Lhx2, as well as Sp5 (Fig. 1A), a direct target gene of the Wnt/β-catenin pathway (Takahashi et al., 2005; Weidinger et al., 2005). Interestingly, Axin2, commonly used as the readout of the canonical Wnt signaling activity, was also expressed at high levels in Placode-I (Fig. 1A).
To analyze Ascl4 expression during the induction of hair and vibrissae development in situ, we performed whole-mount in situ hybridization (WMISH). At E12.5, Ascl4 expression was detected in the whisker placodes (Fig. 1B). By E14.5, Ascl4 mRNA was also localized in the hair placodes of back skin (Fig. 1C) (Sulic et al., 2023). At both developmental stages, Ascl4 expression appeared to be restricted to epithelial compartment, consistent with transcriptomic profiling of E14.5 whole skin by bulk and scRNA-seq (hair-gel.net, Sennett et al., 2015; kasperlab.org/embryonicskin, Jacob et al., 2023).
Fig. 1. Ascl4 is expressed in signaling centers of hair follicles and teeth.
(A) Stacked violin plots showing expression of selected genes in E14.5 hair placode-enriched scRNA-seq dataset (Sulic et al., 2023). Placode-I to Placode-IV clusters represent genuine placode populations, while other clusters represent various interfollicular cell populations, as detailed in Sulic et al., 2023. (B,C) Expression of Ascl4 was analyzed by whole mount in situ hybridization in E12.5 heads (n=5) (B) and E14.5 embryos (n=8) (C). Arrow in (B) indicates expression of Ascl4 in the whisker placodes. Scale bar, 500 µm. (D) Stacked violin plots showing expression of Ascl4, and selected enamel knot markers in the epithelial cell populations of the E14.5 molar tooth scRNA-seq dataset (Hallikas et al., 2021). (E,F) Expression of Ascl4 was analyzed by whole mount in situ hybridization in E12.5 lower jaw (n=7) (E) and E12.5 mammary buds (n=8) (F). Scale bar, 200 µm. Arrows indicate Ascl4 expression in incisor and mammary buds, respectively.
Many genes and pathways have shared functions and similar expression patterns across various ectodermal appendages (Mikkola and Millar, 2006), prompting us to explore Ascl4 expression also in the embryonic tooth and mammary gland. We have recently generated scRNA-seq data on developing molar teeth at E14.5 (Hallikas et al., 2021), a stage characterized by the appearance of the enamel knot, a signaling center governing tooth cusp patterning (Jernvall and Thesleff, 2000). Analysis of Ascl4 expression in this dataset revealed that it was restricted to the epithelium, in a rather small cluster of cells (Fig. S1A). Further sub-clustering of the epithelial cells highlighted Ascl4 enrichment in a cell population defined by high expression of Shh, Wnt10b, Cdkn1a, Fgf20, Dkk4, and Sp5 (Fig. 1D), all known markers of the enamel knot (Vaahtokari et al., 1996; Jernvall et al., 1998; Fliniaux et al., 2008; Porntaveetus et al., 2011; Ahtiainen et al., 2016). This identifies Ascl4 as a novel enamel knot marker. Also Axin2 was enriched in the same cell population (Fig. 1D), consistent with previous studies reporting high Wnt signaling activity in the enamel knot (Liu et al., 2008; Lohi, Tucker and Sharpe, 2010; Ahtiainen et al., 2016). To assess if Ascl4 is expressed already during the tooth placode stage, we performed WMISH at E12.5. At this stage, Ascl4 transcripts were detected in the incisor primordia, albeit at low levels (Fig. 1E). Finally, we explored the expression of Ascl4 in the mammary bud. WMISH revealed weak Ascl4 expression in the mammary bud area at E12.5 (Fig. 1F), specifically in a region surrounding the mammary buds, forming a ring-like pattern.
Ascl4 is a likely transcriptional target of the Wnt/β-catenin pathway
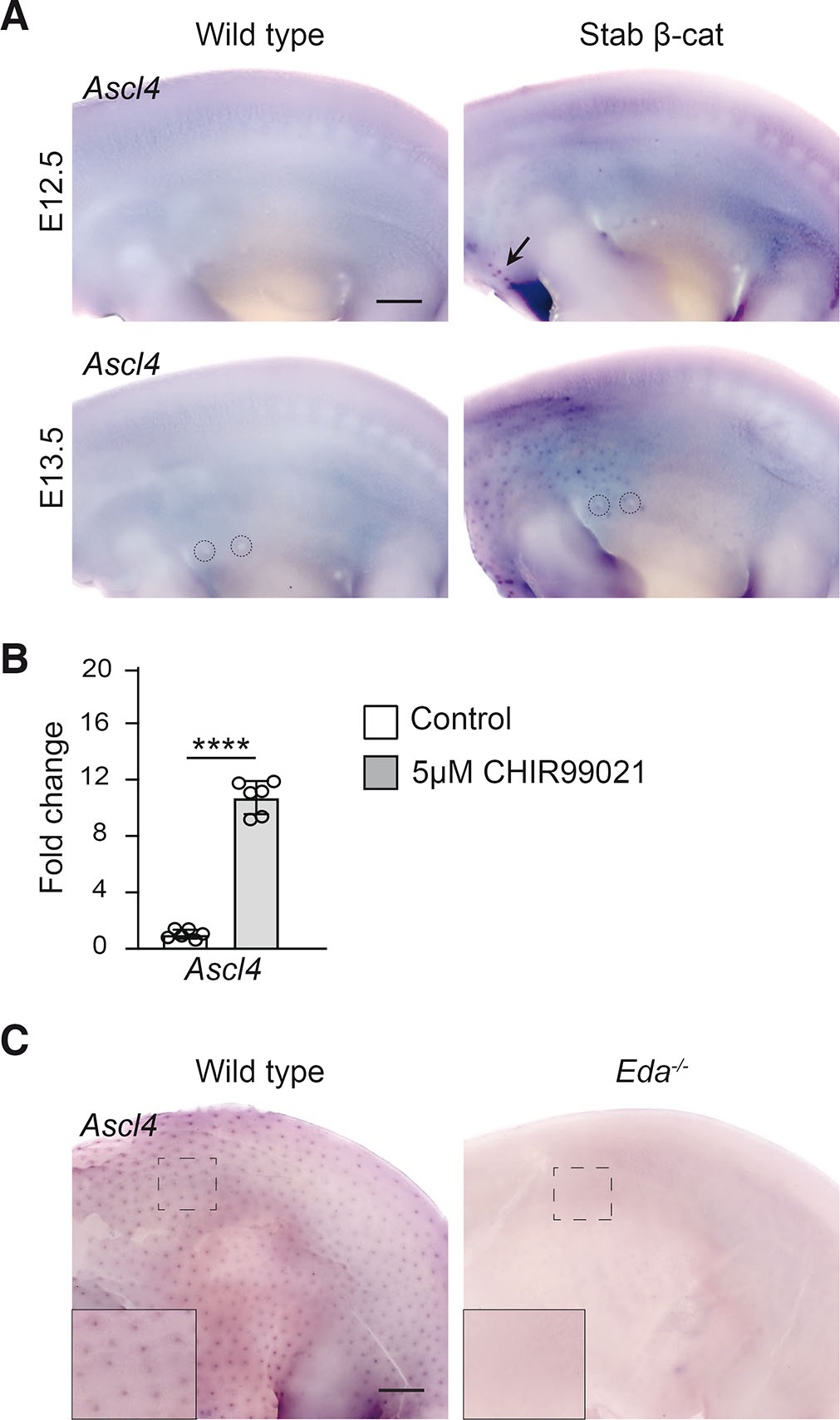
Since Ascl4 expression was detected at sites of high Wnt activity, we next investigated whether it acts downstream of the Wnt pathway. To explore the link between Ascl4 and Wnt signaling in vivo, we analyzed Ascl4 expression in mice with forced epithelial activation of β-catenin (Närhi et al., 2008), expressing one wild-type and one exon 3 deleted allele of Ctnnb1 (encoding a stabilized form of β-catenin) in the mutant skin epithelium. At E12.5, Ascl4 transcripts were detected in the mammary line, as well as in ectopic hair placodes that begin to emerge at this stage (Fig. 2A). A day later, Ascl4 was still undetectable in the back skin of control embryos, whereas its expression had intensified in the ectopic hair placodes and in mammary buds of stab-β-cat embryos. The latter finding is in line with our recent RNA sequencing data that revealed a 7.6-fold upregulation of Ascl4 expression in stab-β-cat mammary buds compared to wild type at E13.5 (Satta et al., 2023).
Fig. 2. Ascl4 is a Wnt target gene.
(A) Ascl4 expression in wild-type (n=3) and stabilized β-catenin (n=3) skin analyzed by whole mount in situ hybridization at E12.5 (top panels) and E13.5 (bottom panels). Arrow indicates the first hair placode-like structures that have appeared at E12.5. Dashed circles highlight mammary buds 2 and 3. Scale bar, 500 μm. (B) qRT-PCR of Ascl4 after 4 hours treatment with 5 µM CHIR99021 or vehicle (n=6). Data are shown as mean ± s.d. ∗∗∗∗p < 0.001. Statistical significance was assessed with two-tailed unpaired Student’s t-test. (C) Ascl4 expression in wild-type (n=4) and Eda-/- embryos (n=4) at E14.5. Insets show higher magnification of the back skin. Scale bar, 500 μm.
These data prompted us to ask whether Ascl4 could be a direct Wnt/β-catenin target gene. To this end, we investigated the ability of CHIR99021, a potent activator of the canonical Wnt pathway (Naujok et al., 2014), to induce Ascl4 expression in E14.5 embryonic back skin explants. Explants were divided into two halves, one treated with vehicle and the other with 5μM CHIR99021, followed by expression analysis by qRT-PCR. 4-hr exposure to CHIR99021 led to a highly significant, 10.7-fold increase in Ascl4 mRNA levels compared to controls (Fig. 2B).
The ectodysplasin (Eda)-Eda receptor (Edar) pathway lies downstream of Wnt signaling in hair placodes (Biggs and Mikkola, 2014). Eda-deficient primary hair placodes are characterized by low Wnt/β-catenin signaling activity and failure to proceed beyond a pre-placode stage (Schmidt-Ullrich et al., 2006; Fliniaux et al., 2008; Zhang et al., 2009). Some of the known Wnt target genes, such as Dkk4 and Fgf20, are expressed in Eda null pre-placodes at E14.5, though at greatly reduced levels (Fliniaux et al., 2008; Huh et al., 2013), suggesting that these genes respond to even low Wnt/β-catenin activity, while many other placode markers such as Shh, and the Wnt target gene Sp5 are undetectable (Huh et al., 2013; Sulic et al., 2023). Similarly, no Ascl4 transcripts could be detected in the back skin of Eda null embryos at E14.5 (Fig. 2C).
Ascl4 is not required for hair follicle morphogenesis
To investigate the function of Ascl4 in ectodermal appendages, we used mice lacking the only exon of Ascl4 resulting in a null allele (hereafter referred to as Ascl4 KO mice). Ascl4 KO mice were viable and fertile, indistinguishable from their wild-type and heterozygous littermates in size, weight, and external appearance (Fig. S1 B-C). Mutant embryos did not exhibit any gross phenotypic differences and were obtained at Mendelian ratios, indicating that the deletion of Ascl4 did not result in embryonic lethality (Fig. S1D).
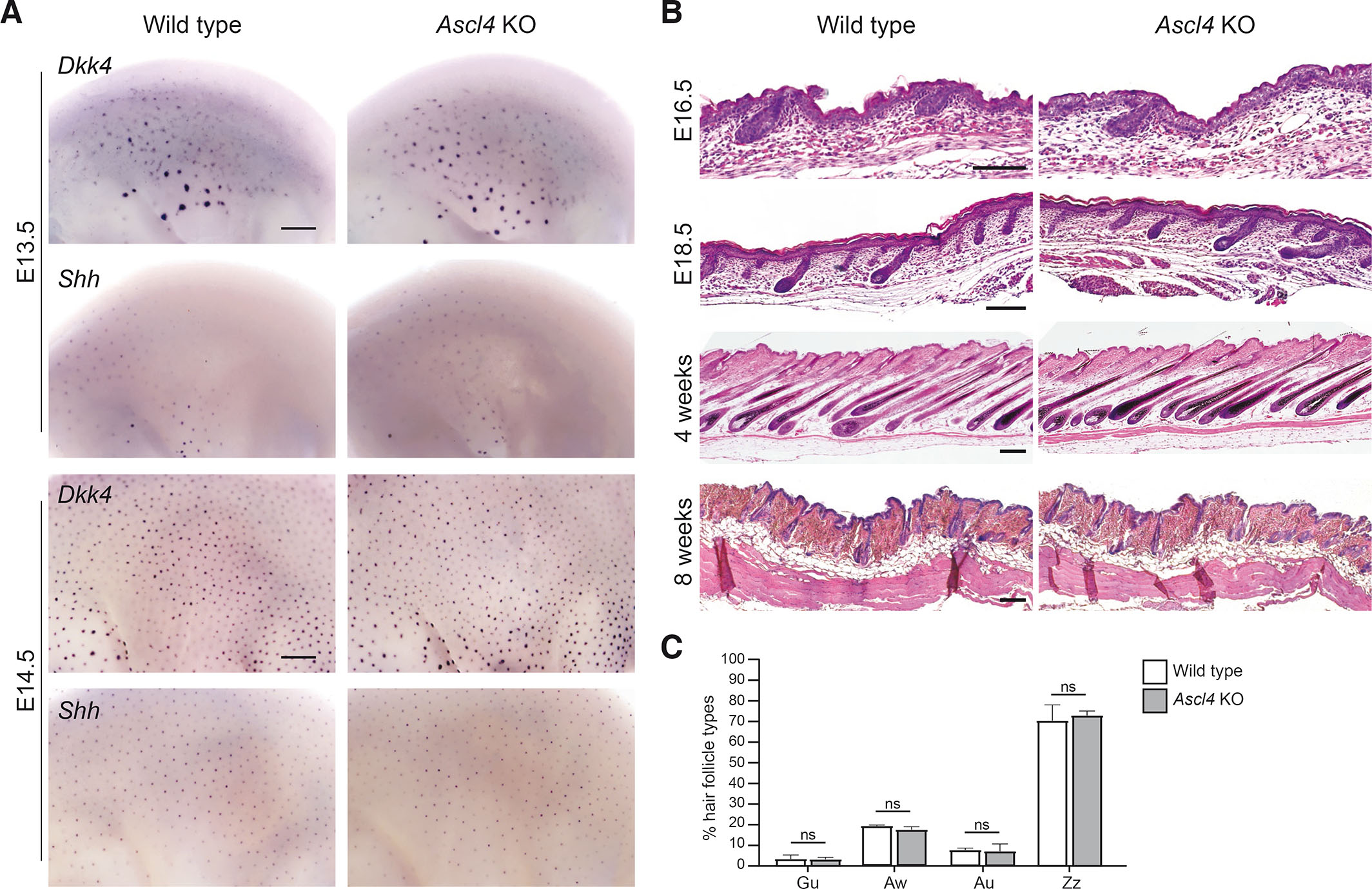
In mice, hair follicles are induced in three successive waves, resulting in the production of four different hair types: the guard (first wave), awl and auchene (second wave), and zigzag (third wave) hairs (Sennett and Rendl, 2012). To assess the functional importance of Ascl4 in hair follicle development, we first examined the expression of three well-characterized hair placode markers – Dkk4, Wnt10b and Shh – in Ascl4 KO embryos using WMISH. At E13.5, Dkk4 expression was readily detectable, while Shh was just starting to emerge, but no difference was observed between the genotypes (Fig. 3A). At E14.5, all genes were expressed in the primary hair placodes of Ascl4 KO embryos, showing spatial and temporal expression comparable to wild-type embryos (Fig. 3A; Fig. S1E). Histological analysis did not reveal any abnormalities in placode morphology either (Fig. S1F). Similar to controls, at E16.5, primary hair follicles in Ascl4 KO embryos had advanced to the peg stage, and secondary hair placodes had formed. By E18.5, all three waves of hair follicles were visible in the mutant skin, with no apparent changes in their sizes, lengths, and distribution (Fig. 3B).
Fig. 3. Ascl4 is not required for hair follicle morphogenesis.
(A) Expression of Dkk4 and Shh was assessed by whole mount in situ hybridization in wild-type (n=4) and Ascl4 KO (n=4) embryos at E13.5 (upper panels) and E14.5 (bottom panels). Scale bar, 500μm. (B) Hematoxylin and eosin staining of wild-type and Ascl4 KO E16.5 (n=6 per genotype), E18.5 (n=6), 4-week-old (n=3), and 8-week-old (n=6) dorsal skin. Scale bar, 100μm. (C) Quantification of hair types in wild-type and Ascl4 KO coat (n=3 per genotype, 500 hairs scored in each mouse) at 8 weeks of age. Statistical significance was assessed with two-tailed unpaired Student’s t-test. ns, not significant; Gu, guard; Aw, awl; Au, auchene; Zz, zigzag
Next, we analyzed whether loss of Ascl4 would impact postnatal hair growth and progression of the hair cycle. Between postnatal day 15-25, hair follicles undergo their first catagen (destruction phase), a short telogen (rest phase) and by 4 weeks of age, the first anagen (growth phase) has commenced (Müller-Röver et al., 2001). At this stage, no morphological differences were observed in the hair follicles of Ascl4 KOs compared to the wild-type littermates (Fig. 3B). At 8 weeks of age, hair follicles had progressed to the second telogen similarly in both genotypes (Fig. 3B). The fur of Ascl4 KO mice contained all four hair types at expected ratios (Fig. 3C) and the external hair structures were not affected either (Fig. S1G). Thus, the loss of Ascl4 does not seem to impair hair follicle development, cycling, or hair shaft formation.
The lack of an obvious hair phenotype in Ascl4 KO mice raised the question whether other Ascl genes could compensate for absence of Ascl4. Our bulk E14.5 RNA-seq data did not reveal the presence of Ascl1 or Ascl3 (sulic.rahtiapp.fi; Sulic et al., 2023), whereas Ascl2 and Ascl5 were detected in the hair placode population, albeit at much lower levels than Ascl4 (Fig. S1H). However, Ascl2 transcripts were enriched in the interfollicular epithelium, not the placode (Fig. S1H). These findings were confirmed by inspection of other published E14.5 databases (Sennett et al., 2015; Jacob et al., 2023). Also postnatally, Ascl2 and Ascl4 were expressed in non-overlapping areas, with Ascl2 being enriched in the interfollicular epidermis, other Ascl genes being expressed at negligible levels in follicular cell populations (Skin-GLOW, https://rstudio-connect.hpc.mssm.edu/skinglow_minerva/).
Ascl4 is not essential for mammary gland and tooth formation
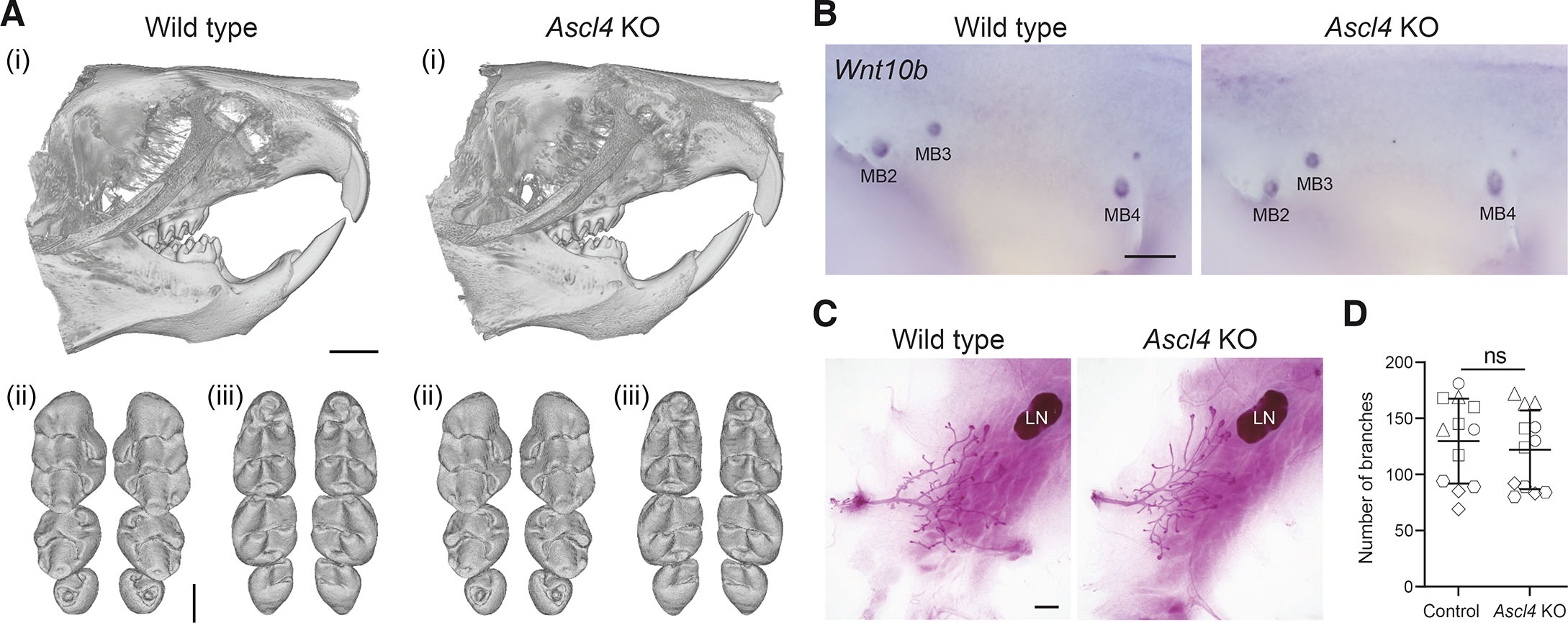
Given that Ascl4 was expressed also in developing teeth and mammary glands, we analyzed the tooth and mammary gland phenotype of Ascl4 KO mice. The dentition of Ascl4 KO mice appeared normal, with molar shapes and sizes indistinguishable from those of wild-type littermate controls (Fig. 4A). To investigate potential redundancy with other family members, we analyzed the expression of Ascl genes in the E14.5 molar tooth scRNA-seq dataset (Hallikas et al., 2021). Of the five family members, only Ascl4 and Ascl5 transcripts could be detected in a clustered manner, while other Ascl genes were expressed only in few sporadic cells (Fig. S1 A,I). Interestingly, Ascl4 and Ascl5 displayed a similar expression pattern, showing enrichment in the enamel knot.
Fig. 4. Ascl4 is dispensable for mammary gland and tooth development.
(A) Representative µCT scans showing incisor (i), and upper (ii) and lower molars (iii) of 4-week-old wild-type (n=6) and Ascl4 KO mice (n=6). Scale bar, 1.5 mm (incisors), and 500 μm (molars). (B) Expression of the mammary bud marker Wnt10b in wild-type (n=5) and Ascl4 KO (n=5) embryos at E12.5. MB, mammary bud. Scale bar, 500 μm. (C) Representative images of whole-mount carmine alum stained 4th mammary gland of 4-week-old wild-type and Ascl4 KO mice. LN, lymph node. Scale bar, 1mm. (D) Quantification of branches (ductal tips) of the 4th mammary gland of control (Ascl4+/+ and Ascl4+/-) (n=12) and Ascl4 KO (n=12) mice at 4 weeks of age. The shape of the symbol indicates littermates. Statistical significance was assessed with two-tailed unpaired Student’s t-test. ns, not significant.
To characterize the Ascl4 KO mammary phenotype, we first analyzed the expression of Wnt10b, a well-known mammary bud marker using WMISH, but found no obvious difference between wild-type and Ascl4 KO embryos at E12.5 (Fig. 4B). The majority of mammary gland growth and branching takes place during puberty, which commences around 3 weeks of age (Macias and Hinck, 2012). The mammary ductal trees of 4-wk old Ascl4 KO mice appeared morphologically indistinguishable from the wild-type littermates (Fig. 4C), a finding confirmed by quantifying the number of ductal tips (Fig. 4D). Ascl4 KO females were also able to nurture their pups (n=6 females, 11 litters, 78 pups), suggesting the absence of any gross lactation defects.
Discussion
The canonical Wnt/β-catenin pathway has long been known to be essential for ectodermal appendage development (Andl et al., 2002; Van Genderen et al., 1994). Here, we identify the transcription factor Ascl4 as a novel target of the Wnt/β-catenin pathway, co-expressed with Shh at signaling centers of developing teeth and hair follicles. Inspection of publicly available RNA-seq hair follicle datasets (accessed via Skin-GLOW, https://rstudio-connect.hpc.mssm.edu/skinglow_minerva/) indicates that also at later developmental stages, Ascl4 is co-expressed with Shh in the transit amplifying cells (activated stem cells) of the hair follicle, as well as the hair matrix encompassing the proliferative precursors giving rise to the hair shaft.
Different levels of Wnt/β-catenin signaling often result in the activation of distinct target genes which in turn determine different cell fates in a context-dependent manner (Matos et al., 2020; Mourao et al., 2021; Söderholm and Cantù, 2021). Our data suggest that Ascl4 expression may require high levels of Wnt/β-catenin activity. Unlike in hair follicles and molar teeth, Ascl4 was not detected in the invaginating mammary bud but rather at its edges. This contrasts with the Wnt target genes and mammary bud markers Dkk4 and Fgf20 (Bazzi et al., 2007; Elo et al., 2017; Fliniaux et al., 2008), which, based on their expression in the Wnt-low milieu of Eda-/- primary hair placodes (Fliniaux et al., 2008; Huh et al., 2013), respond to low levels of Wnt activation. This suggests that the mammary bud displays lower level of Wnt signaling activity than the hair placode or the enamel knot, possibly contributing to the different skin appendage identities. In line with this hypothesis, forced stabilization of β-catenin in the mammary bud led to ectopic expression of Ascl4 along with several other hair placode markers including Shh, as well as Fgf4 (Satta et al., 2023), an enamel knot –specific Wnt target gene (Jernvall et al., 1994; Kratochwil et al., 2002).
The bHLH transcription factors, including the Ascl family, play a central role in cell proliferation, cell fate specification, and differentiation (Ledent and Vervoort, 2001). This, together with the Ascl4 expression pattern data, prompted us to analyze the phenotype of Ascl4 null mice. However, we found no evidence for defective hair follicle morphogenesis, or tooth or mammary gland abnormalities. One possibility is that other Ascl transcription factors compensate for Ascl4. In developing teeth, our scRNA-seq data revealed co-expression of Ascl5 with Ascl4 in the enamel knot, a finding in line with Ascl5 (also known as AmeloD) immunostaining data (Chiba et al., 2019; He et al., 2019). Deficiency in Ascl5 leads to enamel hypoplasia, yet the molar cusp pattern appears relatively normal (Chiba et al., 2019; Jia et al., 2022). We propose this could be due to redundancy with Ascl4. In the hair placodes, Ascl5 and Ascl2 were detected in the hair placodes by RNA-seq. Although both were expressed at low levels, we cannot exclude the possibility that they together, or alone, could function redundantly with Ascl4. An alternative option is that some other bHLH transcription factor compensates for loss of Ascl4. Future studies involving compound mouse mutants are warranted to address these possibilities.
Materials and Methods
Mouse strains
For in situ hybridization experiments, NMRI mice (Envigo) were used, unless stated otherwise. C57BL/6NJ-Ascl4em1(IMPC)J/Mmjax mouse line (MMRRC_#046169-JAX, here referred to as Ascl4 KO), were obtained from the Jackson Laboratory and maintained on C57Bl/6J background. This strain was generated by the Knockout Mouse phenotyping program (KOMP), by deleting the only exon of Ascl4 gene by Crispr-Cas9 technology, resulting in a null allele. Transgenic mouse lines used in this study have been described elsewhere as indicated: Eda null mice (Pispa et al., 1999), Ctnnb1flox3/flox3 (Harada et al., 1999) and K14Cre/wt mice carrying a knock-in of Cre in the Krt14 locus (Huelsken et al., 2001). Embryos were staged according to limb development and other morphological criteria; noon of the vaginal plug was considered as embryonic day (E) 0.5. All mouse studies were approved and carried out in accordance with the guidelines of the Finnish National Experiment Board. Sex of the mice used in the study was not controlled, except only females were used for postnatal mammary gland analysis.
Whole mount in situ hybridization
For whole-mount in situ hybridization (WMISH), embryos were fixed in 4% paraformaldehyde in PBS overnight, and dehydrated using a series of methanol. WMISH was performed manually as previously described (Mustonen et al., 2004). BM Purple AP Substrate precipitating Solution (Roche, Mannheim, Germany) was used to visualize the digoxigenin-labelled riboprobes. The following antisense RNA probes were used: Ascl4 (Sulic et al., 2023), Shh (Echelard et al., 1993), Dkk4 (Fliniaux et al., 2008), and Wnt10b (Wang and Shackleford, 1996). The samples were imaged using Zeiss 456 AxioZoom.V16 stereomicroscope with PlanZ 1.0x/C objective and Axiocam 305 color camera (Zeiss, Oberkochen, Germany), and handled using ZEN 2.3 lite software (Zeiss, Oberkochen, Germany).
Hanging drop culture, and qRT-PCR
The hanging drop culture was performed as previously described (Pummila et al., 2007; Fliniaux et al., 2008). Briefly, E14.5 skins were dissected from NMRI embryos, and cut in half along the dorsal midline: one half was used as a control and the other one treated with CHIR99021. Samples were submerged in DMEM, 10% FBS, 1% penicillin-streptomycin supplemented with either 5 μM CHIR99021 (CT99021) (Sigma-Aldrich, St. Louis, MO) or the respective volume of DMSO as a vehicle control, and placed individually in 50μl hanging drops of under the lid of a 35 mm culture dish containing medium/PBS to prevent evaporation and maintained in a cell culture incubator at 37°C, 5% CO2 for 4 hours. Skin halves were collected directly to TRIzol reagent (Sigma-Aldrich, St. Louis, MO) and stored at -80°C until RNA extraction. Total RNA was extracted using the Direct-zol RNA MicroPrep kit (Zymo Research, Irvine, CA) according to manufacturer’s instructions. cDNA synthesis for qRT-PCR was done with iScript gDNA Clear cDNA Synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions, using 1 μg of RNA as input, and diluted to 10ng/μl.
qRT-PCR was carried out with CFX96TM Real-Time System C1000Touch Thermal Cycler (Bio-Rad, Hercules, CA) using FAST SYBR Green master mix (Thermo Fisher), in triplicate wells. The following cycling protocol was used: Initial denaturation 3 min at 95°C, followed by 45 cycles of denaturation 10 s at 95°C and annealing/extension 50 s at 60°C. qRT-PCR data was analyzed with CFX Manager (Bio-Rad, Hercules, CA) software and fold changes were calculated using the ∆∆Ct method (Livak and Schmittgen, 2001). These were normalized to Hprt gene. qRT-PCR primers were designed as following using NCBI Primer BLAST software: Ascl4 (NM_001163614.2) (Forward primer: TGCTGACCCAAGGAGTGAAT; Reverse primer: CGCCTCATCAACGCTTGTAT), Hprt (NM_013556.2) (Forward primer: CAGTCCCAGCGTCGTGATTA; Reverse primer: TCGAGCAAGTCTTTCAGTCCT).
Histology
For histology, tissues were fixed overnight in freshly prepared 4% paraformaldehyde in phosphate-buffered saline and embedded in paraffin. Transversal sections of 5μm were stained by hematoxylin and eosin using standard protocols and imaged using Axio Imager M.2 widefield microscope equipped with Plan-Neofluar 20x/0.5 objective and AxioCam HRc camera (Zeiss, Oberkochen, Germany) using bright field microscopy.
Analysis of hair types
Hair shafts were collected from 8 week-old mice. Fur was spread onto Whatman filter papers covered in tape, and imaged with Zeiss 456 AxioZoom.V16 stereomicroscope with PlanZ 1.0x/C objective and Axiocam 305 color camera (Zeiss, Oberkochen, Germany). Pictures were handled in ImageJ (Fiji), and individual hair were scored and counted manually.
Micro-Computed Tomography
Head samples were collected from 4-week-old wild-type and Ascl4 KO mice and stored in 70% EtOH. To scan the teeth, heads were immobilized in 1% agarose. X-ray scanning was performed using Bruker 1272 μCT scanner at the μCT laboratory in the Department of Physics, University of Helsinki. The source voltage of 70 kV and source current of 142 μA was used with 0.5-mm aluminum filter. A voxel size of 4.5 μm and 18 μm for molars and incisors were used, respectively. For the reconstruction of the μCT scans, NRecon software (1.7.4.2) with the ring artefact correction of 18 was used. Tooth region was cropped and reconstructed as 8-bit tiff files in Fiji-ImageJ (version 1.53c), and segmentation was done using Avizo (release 9.0.1). For molars, the files were saved as.stl files in ImageJ (Fiji), and final 3D images were obtained using MeshLab (version 2021.10).
Carmine Alum Staining
4-week-old (29 days) female mice were sacrificed, and the entire mammary gland 4 with fat pad was dissected out and spread on glass slides, fixed overnight in acid alcohol (2:8 mixture of glacial acetic acid and 100% ethanol), rehydrated through ethanol series, and stained in carmine alum (Sigma-Aldrich) overnight. Stained tissues were dehydrated in ethanol series, cleared in xylene, and mounted with Depex (BDH). Images were taken with Zeiss 456 AxioZoom.V16 stereomicroscope with PlanZ 1.0x/C objective and Axiocam 305 color camera (Zeiss, Oberkochen, Germany). Number of tips were quantified manually using Image J (Fiji).
scRNA-seq data analysis
E14.5 hair placode-enriched (GSE212674) and E14.5 molar teeth (GSE142201) scRNA-seq datasets were analyzed as previously described (Hallikas et al., 2021; Sulic et al., 2023). Molar data was visualized using t-distributed stochastic neighbor embedding (tSNE) dimension reduction in R package Seurat v4.2.0.1. (Hao et al., 2021), R v4.3.1/RStudio v2023.09.1+494 (Posit Software, PBC). Molar epithelial cells were identified based on expression of Krt14 and Epcam, subsetted and subclustered following Seurat's documentation, using 40 dimensions and resolution of 0.2. Stacked violin plots were produced using scCustomize R package v1.1.3 (Marsh SE, 2021).
Statistical analysis
For qRT-PCR data, quantification of hair types, mammary gland tips, and weight data, statistical analyses were performed using the GraphPad Prism 7 software and two tailed unpaired Student’s T test. p < 0.05 level of confidence was accepted for statistical significance. Accordance to Mendelian genetics was assessed using the GraphPad Prism 7 software and the Chi-square test, with p value <0.05 considered statistically significant.
Supplementary Material
Acknowledgements
We thank Ms. Raija Savolainen for excellent technical assistance, Mr. Otto Mäkelä for support in realizing histology and in situ experiments, and past and present members of the Mikkola and Jernvall labs for stimulating discussions. Mouse studies were carried out with the support of HiLIFE Laboratory Animal Center Core Facility, University of Helsinki. This work was financially supported by the Sigrid Jusélius Foundation (MLM), the Finnish Cultural Foundation (VP), Ella and Georg Ehrnrooth Foundation (JPS), and the doctoral program in Oral Sciences of the University of Helsinki (AKH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
Ahtiainen L., Uski I., Thesleff I., Mikkola M. L. (2016). Early epithelial signaling center governs tooth budding morphogenesis. Journal of Cell Biology 214: 753-767.
Andl T., Reddy S. T., Gaddapara T., Millar S. E. (2002). WNT Signals Are Required for the Initiation of Hair Follicle Development. Developmental Cell 2: 643-653.
Balic A., Thesleff I. (2015). Tissue Interactions Regulating Tooth Development and Renewal. In Craniofacial Development. (Ed. Chai Y., ) Elsevier.
Bazzi H., Fantauzzo K. A., Richardson G. D., Jahoda C. A.B., Christiano A. M. (2007). The Wnt inhibitor, Dickkopf 4, is induced by canonical Wnt signaling during ectodermal appendage morphogenesis. Developmental Biology 305: 498-507.
Biggs L. C., Mikkola M. L. (2014). Early inductive events in ectodermal appendage morphogenesis. Seminars in Cell & Developmental Biology 25-26: 11-21.
Chiba Y., He B., Yoshizaki K., Rhodes C., Ishijima M., Bleck C. K.E., Stempinski E., Chu E. Y., Nakamura T., Iwamoto T., de Vega S., Saito K., Fukumoto S., Yamada Y. (2019). The transcription factor AmeloD stimulates epithelial cell motility essential for tooth morphology. Journal of Biological Chemistry 294: 3406-3418.
Chu E. Y., Hens J., Andl T., Kairo A., Yamaguchi T. P., Brisken C., Glick A., Wysolmerski J. J., Millar S. E. (2004). Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development 131: 4819-4829.
Echelard Y., Epstein D. J., St-Jacques B., Shen L., Mohler J., McMahon J. A., McMahon A. P. (1993). Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75: 1417-1430.
Elo T., Lindfors P. H., Lan Q., Voutilainen M., Trela E., Ohlsson C., Huh S.H., Ornitz D. M., Poutanen M., Howard B. A., Mikkola M. L. (2017). Ectodysplasin target gene Fgf20 regulates mammary bud growth and ductal invasion and branching during puberty. Scientific Reports 7: 5049.
Fliniaux I., Mikkola M. L., Lefebvre S., Thesleff I. (2008). Identification of dkk4 as a target of Eda-A1/Edar pathway reveals an unexpected role of ectodysplasin as inhibitor of Wnt signalling in ectodermal placodes. Developmental Biology 320: 60-71.
García-Bellido A., de Celis J. F. (2009). The Complex Tale of the achaete–scute Complex: A Paradigmatic Case in the Analysis of Gene Organization and Function During Development. Genetics 182: 631-639.
Guillemot F., Hassan B. A. (2017). Beyond proneural: emerging functions and regulations of proneural proteins. Current Opinion in Neurobiology 42: 93-101.
Hallikas O., Das Roy R., Christensen M. M., Renvoisé E., Sulic A.M., Jernvall J. (2021). System‐level analyses of keystone genes required for mammalian tooth development. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution 336: 7-17.
Hao Y., Hao S., Andersen-Nissen E., Mauck W. M., Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R. (2021). Integrated analysis of multimodal single-cell data. Cell 184: 3573-3587.e29.
Harada N. (1999). Intestinal polyposis in mice with a dominant stable mutation of the beta -catenin gene. The EMBO Journal 18: 5931-5942.
He B., Chiba Y., Li H., de Vega S., Tanaka K., Yoshizaki K., Ishijima M., Yuasa K., Ishikawa M., Rhodes C., Sakai K., Zhang P., Fukumoto S., Zhou X., Yamada Y. (2019). Identification of the Novel Tooth-Specific Transcription Factor AmeloD. Journal of Dental Research 98: 234-241.
Huelsken J., Vogel R., Erdmann B., Cotsarelis G., Birchmeier W. (2001). β-Catenin Controls Hair Follicle Morphogenesis and Stem Cell Differentiation in the Skin. Cell 105: 533-545.
Huh S.H., Närhi K., Lindfors P. H., Häärä O., Yang L., Ornitz D. M., Mikkola M. L. (2013). Fgf20 governs formation of primary and secondary dermal condensations in developing hair follicles. Genes & Development 27: 450-458.
Iwafuchi-Doi M., Zaret K. S. (2016). Cell fate control by pioneer transcription factors. Development 143: 1833-1837.
Jacob T., Annusver K., Czarnewski P., Dalessandri T., Kalk C., Levra Levron C., Campamà Sanz N., Kastriti M. E., Mikkola M. L., Rendl M., Lichtenberger B. M., Donati G., Björklund K., Kasper M. (2023). Molecular and spatial landmarks of early mouse skin development. Developmental Cell 58: 2140-2162.e5.
Jernvall J., Åberg T., Kettunen P., Keränen S., Thesleff I. (1998). The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development 125: 161-169.
Jernvall J., Kettunen P., Karavanova I., Martin L., Thesleff I., (1994). Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. International Journal of Developmental Biology 38: 463-469.
Jernvall J., Thesleff I. (2000). Reiterative signaling and patterning during mammalian tooth morphogenesis. Mechanisms of Development 92: 19-29.
Jia L.L., Chiba Y., Saito K., Yoshizaki K., Tian T., Han X., Mizuta K., Chiba M., Wang X., Al Thamin S., Yamada A., Fukumoto S. (2022). The tooth‐specific basic helix‐loop‐helix factor AmeloD promotes differentiation of ameloblasts. Journal of Cellular Physiology 237: 1597-1606.
Jonsson M., Björntorp Mark E., Brantsing C., Brandner J. M., Lindahl A., Asp J. (2004). Hash4, a novel human achaete-scute homologue found in fetal skin. Genomics 84: 859-866.
Järvinen E., Salazar-Ciudad I., Birchmeier W., Taketo M. M., Jernvall J., Thesleff I. (2006). Continuous tooth generation in mouse is induced by activated epithelial Wnt/β-catenin signaling. Proceedings of the National Academy of Sciences 103: 18627-18632.
Kratochwil K., Galceran J., Tontsch S., Roth W., Grosschedl R. (2002). FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1 −/− mice. Genes & Development 16: 3173-3185.
Ledent V., Vervoort M. (2001). The Basic Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Research 11: 754-770.
Lefebvre L. (2012). The placental imprintome and imprinted gene function in the trophoblast glycogen cell lineage. Reproductive BioMedicine Online 25: 44-57.
Liu F., Chu E. Y., Watt B., Zhang Y., Gallant N. M., Andl T., Yang S. H., Lu M.M., Piccolo S., Schmidt-Ullrich R., Taketo M. M., Morrisey E. E., Atit R., Dlugosz A. A., Millar S. E. (2008). Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Developmental Biology 313: 210-224.
Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402-408.
Loh K. M., van Amerongen R., Nusse R. (2016). Generating Cellular Diversity and Spatial Form: Wnt Signaling and the Evolution of Multicellular Animals. Developmental Cell 38: 643-655.
Lohi M., Tucker A. S., Sharpe P. T. (2010). Expression of Axin2 indicates a role for canonical Wnt signaling in development of the crown and root during pre‐ and postnatal tooth development. Developmental Dynamics 239: 160-167.
Macias H., Hinck L. (2012). Mammary gland development. WIREs Developmental Biology 1: 533-557.
Matos I., Asare A., Levorse J., Ouspenskaia T., de la Cruz-Racelis J., Schuhmacher L.N., Fuchs E. (2020). Progenitors oppositely polarize WNT activators and inhibitors to orchestrate tissue development. eLife 9: e54304.
Mikkola M. L., Millar S. E. (2006). The Mammary Bud as a Skin Appendage: Unique and Shared Aspects of Development. Journal of Mammary Gland Biology and Neoplasia 11: 187-203.
Mourao L., Zeeman A.L., Wiese K.E., Bongaarts A., Oudejans L.L., Martinez I.M., Grift Y.B.C., van de Jonkers J., van Amerongen R., (2021). Hyperactive WNT/CTNNB1 signaling induces a competing cell proliferation and epidermal differentiation response in the mouse mammary epithelium. BioRxiv Preprint: 2021.06.22.449461.
Müller-Röver S., Foitzik K., Paus R., Handjiski B., van der Veen C., Eichmüller S., McKay I. A., Stenn K. S. (2001). A Comprehensive Guide for the Accurate Classification of Murine Hair Follicles in Distinct Hair Cycle Stages. Journal of Investigative Dermatology 117: 3-15.
Mustonen T., Ilmonen M., Pummila M., Kangas A. T., Laurikkala J., Jaatinen R., Pispa J., Gaide O., Schneider P., Thesleff I., Mikkola M. L. (2004). Ectodysplasin A1 promotes placodal cell fate during early morphogenesis of ectodermal appendages. Development 131: 4907-4919.
Närhi K., Järvinen E., Birchmeier W., Taketo M. M., Mikkola M. L., Thesleff I. (2008). Sustained epithelial β-catenin activity induces precocious hair development but disrupts hair follicle down-growth and hair shaft formation. Development 135: 1019-1028.
Naujok O., Lentes J., Diekmann U., Davenport C., Lenzen S. (2014). Cytotoxicity and activation of the Wnt/beta-catenin pathway in mouse embryonic stem cells treated with four GSK3 inhibitors. BMC Research Notes 7: 273.
Pispa J., Jung H.S., Jernvall J., Kettunen P., Mustonen T., Tabata M. J., Kere J., Thesleff I. (1999). Cusp Patterning Defect in Tabby Mouse Teeth and Its Partial Rescue by FGF. Developmental Biology 216: 521-534.
Porntaveetus T., Otsuka-Tanaka Y., Basson M. A., Moon A. M., Sharpe P. T., Ohazama A. (2011). Expression of fibroblast growth factors (Fgfs) in murine tooth development. Journal of Anatomy 218: 534-543.
Pummila M., Fliniaux I., Jaatinen R., James M. J., Laurikkala J., Schneider P., Thesleff I., Mikkola M. L. (2007). Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development 134: 117-125.
Satta J. P., Lan Q., Taketo M. M., Mikkola M. L. (2023). Stabilization of Epithelial β-Catenin Compromises Mammary Cell Fate Acquisition and Branching Morphogenesis. Journal of Investigative Dermatology In Press: S0022-202X(23)03201-3.
Schmidt-Ullrich R., Tobin D. J., Lenhard D., Schneider P., Paus R., Scheidereit C. (2006). NF-κB transmits Eda A1/EdaR signalling to activate Shh and cyclin D1 expression, and controls post-initiation hair placode down growth. Development 133: 1045-1057.
Sennett R., Rendl M. (2012). Mesenchymal–epithelial interactions during hair follicle morphogenesis and cycling. Seminars in Cell & Developmental Biology 23: 917-927.
Sennett R., Wang Z., Rezza A., Grisanti L., Roitershtein N., Sicchio C., Mok K. W., Heitman N. J., Clavel C., Ma’ayan A., Rendl M. (2015). An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Developmental Cell 34: 577-591.
Söderholm S., Cantù C. (2021). The WNT/β‐catenin dependent transcription: A tissue‐specific business. WIREs Mechanisms of Disease 13: e1511.
Spina E., Cowin P. (2021). Embryonic mammary gland development. Seminars in Cell & Developmental Biology 114: 83-92.
Sulic A.M., Das Roy R., Papagno V., Lan Q., Saikkonen R., Jernvall J., Thesleff I., Mikkola M. L. (2023). Transcriptomic landscape of early hair follicle and epidermal development. Cell Reports 42: 112643.
Takahashi M., Nakamura Y., Obama K., Furukawa Y., (2005). Identification of SP5 as a downstream gene of the beta-catenin/Tcf pathway and its enhanced expression in human colon cancer. International journal of oncology 27: 1483-1487.
Tomann P., Paus R., Millar S. E., Scheidereit C., Schmidt-Ullrich R. (2016). LHX2 is a direct NF-κB target gene that promotes primary hair follicle placode down-growth. Development 143: 1512-1522.
Vaahtokari A., Åberg T., Jernvall J., Keränen S., Thesleff I. (1996). The enamel knot as a signaling center in the developing mouse tooth. Mechanisms of Development 54: 39-43.
van der Flier L. G., van Gijn M. E., Hatzis P., Kujala P., Haegebarth A., Stange D. E., Begthel H., van den Born M., Guryev V., Oving I., van Es J. H., Barker N., Peters P. J., van de Wetering M., Clevers H. (2009). Transcription Factor Achaete Scute-Like 2 Controls Intestinal Stem Cell Fate. Cell 136: 903-912.
van Genderen C., Okamura R. M., Fariñas I., Quo R. G., Parslow T. G., Bruhn L., Grosschedl R. (1994). Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice.. Genes & Development 8: 2691-2703.
Wang J., Shackleford G. M., (1996). Murine Wnt10a and Wnt10b: cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene 13: 1537-1544.
Weidinger G., Thorpe C. J., Wuennenberg-Stapleton K., Ngai J., Moon R. T. (2005). The Sp1-Related Transcription Factors sp5 and sp5-like Act Downstream of Wnt/β-Catenin Signaling in Mesoderm and Neuroectoderm Patterning. Current Biology 15: 489-500.
Yu T., Klein O. D. (2020). Molecular and cellular mechanisms of tooth development, homeostasis and repair. Development 147: 184754.
Zhang Y., Andl T., Yang S. H., Teta M., Liu F., Seykora J. T., Tobias J. W., Piccolo S., Schmidt-Ullrich R., Nagy A., Taketo M. M., Dlugosz A. A., Millar S. E. (2008). Activation of β-catenin signaling programs embryonic epidermis to hair follicle fate. Development 135: 2161-2172.
Zhang Y., Tomann P., Andl T., Gallant N. M., Huelsken J., Jerchow B., Birchmeier W., Paus R., Piccolo S., Mikkola M. L., Morrisey E. E., Overbeek P. A., Scheidereit C., Millar S. E., Schmidt-Ullrich R. (2009). Reciprocal Requirements for EDA/EDAR/NF-κB and Wnt/β-Catenin Signaling Pathways in Hair Follicle Induction. Developmental Cell 17: 49-61.