Int. J. Dev. Biol. 68: 199 - 209 (2024)
Special Issue: Developmental Biology in Nordic Countries
Epigenetic and transcriptional regulation of neuron phenotype
Open Access | Review | Published: 21 August 2024
Abstract
Understanding the structure and function of cells is central to cell biology and physiology. The ability to control cell function may benefit biomedicine, such as cell-replacement therapy or regeneration. If structure defines function and cells are composed of water, lipids, small metabolites, nucleic acids, and proteins, of which the latter are largely encoded by the DNA present in the same cell, then one may assume that the cell types and variation in cellular phenotypes are shaped by differential gene expression. Cells of the same cell type maintain a similar composition. In this review, I will discuss the epigenetic and transcription regulation mechanisms guiding cell fate- specific gene expression in developing neural cells. Differentiation involves processes of cell-fate selection, commitment and maturation, which are not necessarily coupled.
Keywords
phenotypic convergence, differentiation, cellular competence, epigenetics, cell fate decision
Regulation of developmental fate transitions
Cell type-specific genome and gene-expression state is largely established by sequence-specific transcription factors (TFs) and local epigenetic modifications. Germ layer identity may be constituted by global editing. For example, preventing methylation at H3K27 via knockout of polycomb group genes Eed and Suz12 shifts the fate of mouse embryonic stem cells (mESC) towards mesendoderm at the expense of ectodermal fate (Yu et al., 2023). However, further diversification into cell types and differentiation is clearly regulated by highly specific TF-DNA interactions.
Cell fate selection is closely associated with specific TFs called “pioneer factors”. Pioneer TFs bind DNA irrespective of its pre-existing conformational state or associated histone modifications and prime chromatin decondensation (Soufi et al., 2012). As such, the pioneer TFs reshape the chromatin landscape, opening new chromatin regions and enabling a new transcriptional state. The ability to bind heterochromatin and the preference for open or closed chromatin varies between TFs. Perhaps only a subset of target enhancers may mediate the pioneer function, while others require chromatin decondensation by prior activators or a topological shift. Little is known about the regulation of topological shifts. Euchromatin and heterochromatin are localized in distinct nuclear compartments, and developmental stage-specific gene enhancers can move between the nuclear compartment cell type specifically (Norrie et al., 2019). Interestingly, generation of induced pluripotent cells can be greatly facilitated by deforming the nucleus (Song et al., 2022).
Together with TFs, the chromatin state at enhancers and promoter CpG islands represent the other side of the coin in successful expression of required genes. Unlike differentiated cells, where a stable state of chromatin and histone modifications are maintained, the epigenetic state is dynamic during development. Developmental gene enhancers are associated with a poised state: a bivalent modification of histones (reviewed in Macrae et al., 2023) (Rada-Iglesias et al., 2011, 2012). Stabilization of the current chromatin state may signify cell-fate commitment.
Gene expression programs and chromatin state in neurons
The nervous system in mammals and vertebrates is remarkably complex in terms of cell type number. Consistent with the higher number of distinct cell types, regulation of gene expression may be more multivariate in the nervous system compared with other tissues. The intergenic regions flanking neural genes have longer intergenic regions (2 times longer than the intergenic regions of tissue-specific genes of other tissues) that also contain more regulatory elements (REs, also known as enhancers and silencers) (Jaura et al., 2022). This feature has emerged in vertebrates, as neural genes in Drosophila and Caenorhabditis elegans are not associated with longer intergenic regions. Neural gene enhancers also seem to be used in a highly cell type-specific manner; in any region of the cortex, the number of region-specific REs vastly exceeds the number of REs active in all regions. For example, different genomic elements are accessible near commonly expressed genes Sst and Negr1 in the cortex, hippocampus, and motor neurons in mice (Jaura et al., 2022).
Interestingly, the neuronal gene expression program in its common (or pan-neuronal) units can be induced by several distinct TF combinations convergently. Mouse embryonic fibroblasts (MEFs) can be reprogrammed in vitro to neurons (iN) using a combination of the TFs Pou3f2 (Brn2), Ascl1, and Myt1l (BAM), or, less efficiently, using Ascl1 alone (Pang et al., 2011). Similarly, expression of the basic helix-loop-helix transcription factor (bHLH TF) Neurog2 can be used to generate iN of glutamatergic identity (Thoma et al., 2012). Ascl1 and Neurog2 function as pioneer factors; however, MEF-derived iN can also be created independent of Ascl1 by coexpression of Sox8 and Mytl1 or Dlx3 and Mytl1 (Wapinski et al., 2013, 2017). The differentiation process is possibly relayed along the regulatory network and may also be activated by downstream layer regulators. Consistent with the hypothesis of feed-forward relay, in Xenopus embryo and mouse P19 cells, a similar glutamatergic neuron differentiation program can be initiated by either Neurog2 or NeuroD4, a direct transcriptional target of Neurog2 (Seo et al., 2007).
The activity of reprogramming factors is guided by the chromatin landscape, but also shapes it. In endogenous neural progenitor cells (NPCs) and MEFs alike, the Ascl1-binding “E-box” motifs are associated with histone marks H3K4me1, H3K27ac, H3K9me3, forming a closed but permissive chromatin state (Wapinski et al., 2013, 2017), while Pou3f2 binding occurs at already open chromatin (Wapinski et al., 2013). In NS5 cell-derived iN, Ascl1 binding to DNA is globally activating in function and binding was preferentially detected in enhancers associated with H3K27ac and H3K4me1, with the proportion of open-chromatin binding increasing during differentiation; 80% of Ascl1-bound sites overlap with DNase I hypersensitivity sites (DHS) in early and 91% in the late differentiation stage. In the genomic loci of NeuroD4, Ap3b2, Mcf2l, and Nrxn3, Ascl1 binding was detected in closed chromatin in the early differentiation stage and precedes the appearance of DHS at binding site (Raposo et al., 2015). In mESC-derived motor neurons, Neurog2 acts as a pioneer factor and the accessible REs are later bound by Pou3f2, Ebf2, Onecut2, and Isl1 (Rhee et al., 2016), where Isl1 is a selector gene in motor neurons. Interestingly, Isl1 first binds DNA broadly, followed by a shift in its genome occupancy, mediated by Onecut2. Onecut2 recruits Isl1, displacing it from "transient" enhancers by protein-protein interaction (Rhee et al., 2016). The function of very short-lived transient enhancers is still obscure. Such binding events might simply immobilize and accumulate TFs.
Altogether, neuronal fate acquisition and differentiation can be envisioned as a two-step process as follows: 1, broad marking and opening of the common neuronal program by pioneer type TFs, and 2, the restriction of a genetic program by cell type-specific factors (selector TFs) that maintain a subset of open chromatin elements while others are silenced.
As several TF combinations can convert mouse and human fibroblasts to iN, it would be interesting to show how the neurogenic programming factors guide cells. This guidance involves both activation of new chromatin regions and chromatin silencing. In iN, Myt1l functions as a lineage-specific repressor. Myt1l is also expressed exclusively in neural tissues in vivo and targets myogenic, cartilage, heart, and lung development-associated genes while neuronal gene promoters are depleted of Myt1l-binding motifs (Mall et al., 2017). The activating neurogenic factor function seems to converge on a “core neuron transcriptome” that includes transcriptional repressors such as RE1-silencing transcription factor Rest and common and specific activators, including the common activator Mecp2, methyl-CpG binding protein 2 (Tsunemoto et al., 2018). Pan-neuronal genes such as Mapt, Tubb3, Map3, and Snap25 are reliably induced in iN generated using different TF combinations (Tsunemoto et al., 2018). However, the genes governing neurotransmitter identity are often not expressed at endogenous levels or are not segregated in iN. For example, BAM-iN cells express both Gad1/2 and Slc17a6 and are excitatory interneurons by function (Pang et al., 2011). Induced pluripotent stem cells (iPSC)-derived Neurog2-iN cultures contain a mixture of molecularly distinct central nervous system (CNS) and peripheral nervous system (PNS) neurons, including motor neurons and forebrain cholinergic and glutamatergic neurons (Chen et al., 2020; Lin et al., 2021).
Obtaining the mature neuron phenotype involves several developmental transitions in vivo. Consistently, mimicking endogenous developmental gene expression in vitro can increase the differentiation efficiency and the homogeneity of the induced neurons. Such guided differentiation assays have been developed for several neurodegenerative disease-associated cell types, including dopaminergic and serotonergic neurons, motor neurons, and various types of GABAergic and glutamatergic neurons (reviewed in Limone et al., 2022). Using this strategy, sequential expression of Sox2 and Foxg1 followed by Ascl1, Dlx5, and Lhx6 has been used to produce cortical interneuron-like cells from mouse and human fibroblasts and human iPSC (Colasante et al., 2015). The in vitro generated cortical GABAergic neurons expressed Arx, Dlx1, Dlx2, Sox6, Satb1, and ErbB4, acquired GABAergic phenotype and morphology, and integrated to mouse hippocampus upon transplantation, forming GABAergic inhibitory synapses in the host tissue (Colasante et al., 2015). The purity and cell-type composition of the iN cultures may need further investigation. Ectopic expression of Dlx5, Lhx6, and phosphorylation-resistant Ascl1 in human pluripotent stem cells can convert 70-90% of cells to Gad1-positive GABAergic neurons. Adding micro-RNAs miR-9 and miR-124 further supported neuronal differentiation (Sun et al., 2016). Molecular marker analysis showed that the protocol used in Sun et al., rather induces a lineage identity and not a cell type, as a mixture of medial ganglionic eminence-derived interneuron subtypes including the SST-, calbindin-, calretinin-, and NPY-expressing interneurons were found. Nevertheless, the transplanted neurons successfully integrated in the mouse cortex, maintaining their subtype identity (Sun et al., 2016). Guided differentiation assays for other neuron types exist. A combination of ectopic Neurog2 with regional cues (retinoic acid and Smoothened inhibitor) and growth factors (GDNF, BDNF, and CNTF) enabled 95% purity of induced lower motor neurons, and the neurons formed neuromuscular connections in vitro (Limone et al., 2023). Arguably, the requirement for transfections or viral transductions is a risk. Production of midbrain dopaminergic neurons from human iPSC requires application of Shh activators, FGF8, and BMP inhibitors, followed by sorting of Corin+ floor plate-like cells and propagation under growth factors (Doi et al., 2014). In vitro differentiated dopaminergic cells have therapeutic potential, as transplantation of these cells into the striatum results in improvement of movement in animal models of Parkinson’s disease (Liu et al., 2012; Kikuchi et al., 2017).
Regulation of the common target genes of basic helix-loop-helix transcription factors
The transcription factors that induce neurogenesis in vivo are called “proneural”. Proneural TFs are expressed in neuronal progenitors and induce differentiation by repressing re-entry to the cell cycle and promoting cell cycle exit. In mouse neuronal precursors, the main proneural factors are the bHLH TFs Ascl1, Neurog2, and Neurog1. bHLH TFs have pioneer TF function (Zhu et al., 2018) and, as discussed earlier, Ascl1 and Neurog2 also promote neuronal fate in vitro.
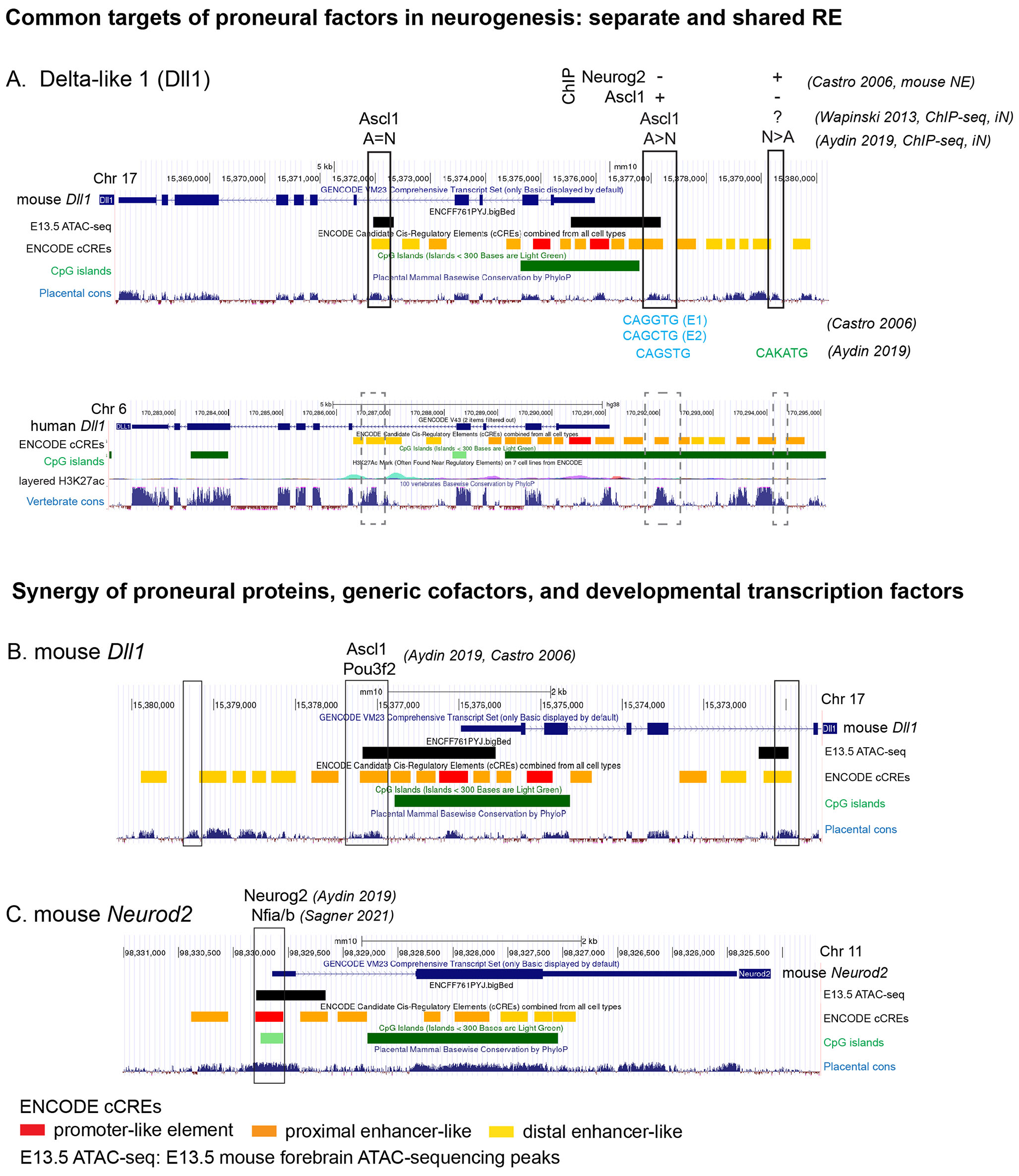
Looking at the global chromatin landscape and the DNA binding activity of Ascl1 or Neurog2 during induced neurogenesis, Ascl1 and Neurog2 were shown to preferentially bind distinct genomic locations. A smaller number of REs could be bound by both TF. These REs contain either multiple E-box sequences or a single binding site, whose sequence is an average of Ascl1-only and Neurog2-only motifs and is probably recognized by both TFs (Fig. 1A) (Aydin et al., 2019). Despite differences in binding locations, Ascl1 and Neurog2 activate somewhat overlapping sets of target genes. The target-gene activation seems to be remarkably cell type dependent, as approximately 80% of common targets were reported in in vitro differentiated mESCs (Aydin et al., 2019) but only 3.1% in reprogrammed astrocytes (Masserdotti et al., 2015). The high number of shared targets would not be surprising as Ascl1 and Neurog2 proteins are structurally and functionally similar. The same binding sites could be used without competition or loss of affinity, as Ascl1 and Neurog2 are rarely coexpressed in developing embryos. However, the situation seems to be more complex. When specific target genes are considered, there are examples of both shared and distinct enhancers mediating proneural function (Fig. 1).
Fig. 1. Convergence of transcription factor activity on the regulatory elements upstream of the Dll1 and Neurod2 genes in developing neuronal precursors.
(A) Genomic loci of the mouse and human Delta-like 1 (Dll1) genes. The conserved regulatory elements (RE) are outlined in boxes. The binding of Ascl1 and Neurog2 in each RE, according to different studies, is shown above the RE. The consensus sequence of the E-box in the RE is shown below the box, when known, according to Castro et al.,2006 and Aydin et al.,2019. A, Ascl1; N, Neurog2; The “=”, “<” and “>” signs indicate whether the RE is preferentially bound by Ascl1, Neurog2 or both, according to Aydin et al.,2019). (B,C) Genomic loci of the mouse Delta-like 1 (Dll1) and Neurod2 genes, showing the synergistic activity of transcription factors in REs. The references are indicated. Mouse genome mm10 or human genome hg38, gene models, location of ENCODE CREs, CpG islands and the conservation in placental or vertebrate animals was fetched from the UCSC genome browser. The E13.5 mouse ATAC-seq features are from the E13.5 mouse forebrain ATAC-seq experiment (ENCODE accession number ENCFF761PYJ).
An important feature of the neurogenic gene expression program induced by proneural factors is the expression of Delta-like genes. Dll1 (Delta-like1) is a common target of Ascl1 and Neurog2, which are thought to act via distinct enhancers (DeltaM and DeltaN, as described in mice and zebrafish) (Castro et al., 2006). The DeltaM enhancer is conserved in vertebrates and contains 2 E-boxes and a POU-HD binding octamer sequence (Fig. 1A). Binding at DeltaM, Ascl1 can activate Dll1 expression alone or synergistically with Pou3f2 (Brn2) (Fig. 1B) (Castro et al., 2006). POU TFs are common cofactors of Ascl1 in CNS. A conserved POU/E-box sequence combination similar to the Dll1 enhancer is found in the proximity of several neurogenesis-associated genes, which could be interpreted as a coregulation unit. These genes include Notch signalling pathway proteins, TFs, neuronal migration regulators, and cell-cycle regulators (Castro et al., 2006). A recent study (Aydin et al., 2019) confirms the preferential binding of Ascl1 to DeltaM and Neurog2 to DeltaN in iN (Fig. 1A). Distinct and shared enhancers for Ascl1 and Neurog2 are also a feature of the mouse Dll3 promoter. The Dll3 promoter contains 7 E-box motifs (E0-E6) that mediate Dll3 gene activation, binding either Ascl1 or Neurog2 alone or synergistically with Tcfe2a and Nhlh1. Interestingly, the E0 motif is critical for Dll3 expression, but the TFs interacting with E0 are unknown and do not include Ascl1, Neurog2, Nhlh1, and Tcfe2a-E12 (Henke et al., 2009).
Another common target of Ascl1 and Neurog2 is Gadd45ɣ (Seo et al., 2007; Huang et al., 2010). Gadd45 proteins are small 18-kDa cytoplasmic and nuclear proteins expressed in postmitotic neuronal cells (Tamura et al., 2012). Gadd45ɣ function is important for G2/M checkpoint, cell cycle withdrawal, and induction of differentiation. Similar to Ascl1 overexpression, Gadd45ɣ overexpression in P19 cells leads to neuronal morphology, physiological characteristics of neurons, and expression of neuronal genes Slc17a6, Syt4, Npy, and Tuj. The Gadd45ɣ promoter contains 4 E-box motifs conserved in mouse, zebrafish, and Xenopus. Its regulation is independent of Pou3f2 function, as HD octamer is not present. Gadd45ɣ overexpression does not induce Ascl1 or Neurog2 expression (Huang et al., 2010). As proneural proteins are downregulated soon after cell cycle exit, Gadd45ɣ may function as a relay protein in the neurogenic program (Table 1).
Table 1
Cell cycle exit regulators, chromatin-, histone-, and DNA-modifying enzymes, and transcription factors expressed in neurons during differentiation
| Developmental stage | DNA-binding domain | Target sequence motif | Human neurological conditions associated with gene mutation | Functional study of human gene mutation in human [HS] or mouse [MM] | |||||
|---|---|---|---|---|---|---|---|---|---|
| “early” | “mid” | “late” | |||||||
| Diff. state | Cell-cycle exit regulators | Lin28a Dnajc2 Dll1 Dll3 Fbxw7 Cdc25b |
Gadd45g | Brain malformations (DLL1) (Fischer-Zirnsak et al., 2019). | |||||
| Chromatin state:generic cofactors | Chromatin modifiers | Hmga1 Hmga2 Hmgb1 Mecp2 |
Hmga1 Hmgb1 Dnmt3b |
HMG-box | structure (HMGA, HMGB) me-CpG (Mecp2) |
Rett syndrome (MECP2) (Collins and Neul, 2022; Good et al., 2021). | |||
| Transcription factors | SOX | Sox11 | Sox2 Sox6 Sox4 |
Sox9 | HMG-box | CTTTGT | Coffin-Siris syndrome (SOX11)(Wang et al., 2023; Al-Jawahiri et al., 2022) Eye malformation (SOX2)(Kelberman et al., 2008; Zenteno et al., 2006) Intellectual disability (SOX4)(Angelozzi et al., 2022; Grosse et al., 2023) |
||
| HLH (inhibitory) | Id3 | Id4 | Id4 | - | - | NA | |||
| bHLH | Tcf12 | Tcf4 | Tcf4 Neurod2 Neurod6 |
basic domain | CANNTG | Pitt-Hopkins syndrome (TCF4)(Popp et al., 2022) Craniosynostosis, neurodevelopmental delay (TCF12)(Kennedy-Williams et al., 2021; Sharma et al., 2013) Neurodevelopmental delay, epileptic encephalopathy (NEUROD2)(Sega et al., 2019) Downregulated in Alzheimer's disease brain (NEUROD6)(Fowler et al., 2015; Satoh et al., 2014) |
TCF4 mutations affect neuron type and differentiation in human cerebral organoids [HS](Papes et al., 2022) Exonic polymorphisms in NEUROD2 associated with differential response to psychoactive drugs [HS](Spellmann et al., 2017) |
||
| NFI | Nfia Nfib |
Nfia Nfib Nfix |
CAAT-box | (T)GCCA(A) | Brain malformations (NFIA)(Negishi et al., 2015; Labonne et al., 2016) Intellectual disability and macrocephaly (NFIB)(Schanze et al., 2018) Marshall-Smith syndrome (NFIX)(Uzman et al., 2023) |
||||
| NR | Nr6a1 (Gcnf) | Nr1a1 (Thra) | cysteine-rich domain (Zn-finger) |
TF-specific sequences | Broad range of neurodevelopmental defects (MYT1L)(Coursimault et al., 2022) Pain insensitivity (ZFXH2)(Habib et al., 2018) Intellectual disabilities (NR1A1)(Krieger et al., 2019) Primrose syndrome (ZBTB20)(Melis et al., 2020) Motor discoordination and apraxia of speech (BCL11A)(Bruce et al., 2022) |
NR6A1 regulates hypocretin transcription in hypothalamus [HS](Tanaka et al., 2010) NR1A1 regulates neurogenesis [HS][MM](Krieger et al., 2019) Bcl11a regulates development of cortical projection neurons and midbrain dopaminergic neurons [MM](Dias et al., 2016; Woodworth et al., 2016; Tolve et al., 2021) |
|||
| ZF | Myt1l | Zfhx2 Zfhx3 Zfhx4 Zfp422 |
Sall3 Zbtb20 Bcl11a |
||||||
| HD | Pou3f2 | Pou2f2 | helix-turn-helix | ATGCAAAT | Neurodevelopmental delay, autism (POU3F2)(Schönauer et al., 2023) | POU3F2 regulates proliferation and differentiation of human NPCs [HS](Chen et al., 2018) | |||
| HD-CUT | Onecut2 | Onecut3 | homeo-domain | ATC[A/G]ATA | NA | ONECUT2 regulates MITF, microphthalmia gene [MM][HS](Jacquemin et al., 2001) | |||
In conclusion, Ascl1 and Neurog2 both function as pioneer TFs, modifying the chromatin accessibility landscape and as cell fate determinants that regulate the terminal differentiation genes. The neuronal differentiation program is somewhat convergent, while the potential to execute the full target program is context-dependent and likely shaped by epigenetic modifications or preset nuclear environment. Proneural TFs can act via shared or unique REs, while the convergently regulated, common target genes have multiple REs.
Phenotypic convergence in invertebrates
In this section, I highlight some interesting recent studies that have addressed the generalizable regulatory logic of cell fate determination. There are exceptional cases where cell fate is defined by a single TF. In the ventral nerve cord motor neurons of the nematode C. elegans, synaptic acetylcholine production as well as acetylcholine neurotransmission genes are activated by COE-type TF unc-3 (Kratsios et al., 2012). In mouse serotonergic neurons, the capacity of serotonin neurotransmission is induced and maintained by the ETS (E26 transformation-specific) family TF Fev (Liu et al., 2010; Wyler et al., 2016). Such critical fate-determining TFs have been called “selector genes”. In most cases, however, the regulatory landscape is complex (Kutejova et al., 2016) and is perhaps better exemplified by the regulation of eat-4/Vglut gene expression in C. elegans glutamatergic neurons, where eat-4 is controlled by a modular enhancer responsive to multiple alternative TFs of different TF classes (Serrano-Saiz et al., 2013). Similar regulatory logic seems to apply for pan-neuronal genes Rab-3, ric-4/Snap25, and snb-1/Vamp, where any partial deletion of an enhancer does not eliminate the expression in all neurons (Stefanakis, Carrera and Hobert, 2015).
A recent study of selector gene-based regulation of cellular phenotype used annotated regulatory links between the known selector genes and terminal differentiation genes in C. elegans for regulatory link modelling (Mora-Martinez, 2021). The model assigned different numbers of selector-type TFs per target gene, depending on the level of cell-type specificity of the target gene: the cell type-specific genes were strongly linked to only 2-5 TFs, while the broadly expressed genes could be equally linked to about 10 different TFs (Mora-Martinez, 2021). In another study, the cell-type markers and expressed TF combinations were correlated in 52 single-cell clusters and 17 cell types isolated from the developing eye of Drosophila melanogaster, using single-cell and bulk RNA-sequencing (Konstantinides et al., 2018). There, 1/3 of the expressed genes correlated with a single TF expression and were expressed on average in 2.2 clusters. The other 2/3 of genes correlated with a combination of TFs and were expressed on average in 22 clusters (Konstantinides et al., 2018). Testing various models of regulation, the expression pattern of neural differentiation genes was better explained by a model where a gene can be regulated by several TFs convergently.
Some common principles arise from these studies. First, only a minority of differentiation genes are expressed in a unique single-cell cluster (or cell type), while the expression of most genes is not highly restricted across cells. Second, different regulatory networks may explain the regulation of more restricted, cell subtype-specific genes and more broadly expressed, cell type- or cell class (pan-neuronal)-specific genes. The model of a redundant modular architecture seems to be common to enhancers regulating the pan-neuronal or neuron class-specific genes, and a master regulator type control (or selector gene model) to the subtype-specific gene expression (Hobert and Kratsios, 2019).
Phenotypic convergence in vertebrate nervous system
Similar to invertebrates, a shared cell lineage or embryonic field of origin does not always correlate with chromatin or gene expression state similarity in vertebrates. Similar molecular identity can be derived from distinct lineages, and these divergences and convergences are also apparent in single-cell RNA-seq using genetic lineage tracers in mice and zebrafish in vivo (Wagner et al., 2018; Chan et al., 2019). However, the genetic regulatory mechanisms are much less studied in vertebrates.
As an example of phenotypic convergence at cell subtype level, inhibitory GABAergic interneuron fate is controlled by distinct selector genes in spatiotemporally and molecularly distinct neuroepithelial cell lineages, such as HD-TFs Dlx1, Dlx2, and Dlx5 in the telencephalon and anterior diencephalon; zinc-finger TF Gata2 and bHLH TF Tal2 in posterior diencephalon and midbrain; and Gata2, Gata3, and Tal1 in hindbrain (Achim, Salminen and Partanen, 2014). bHLH TF Ptf1a acts as a selector gene in GABAergic dorsal spinal cord interneurons, cerebellar granule neurons, and GABA- or glycinergic retinal amacrine cells (Jin and Xiang, 2019). All these TFs function as selector genes, promoting GABAergic over glutamatergic neurotransmitter phenotype. Interestingly, Ptf1a also promotes differentiation of the acinar cells of the pancreas (where it regulates amino acid biosynthesis and secretion) and the paracrine serotonergic neurons in the enteric nervous system (Hoang et al., 2016; Jin and Xiang, 2019). In neurons, GABAergic neurotransmission is governed by the expression of glutamic acid decarboxylase genes Gad1 and Gad2 and the GABA vesicular transporter gene Slc32a1, which are expressed in all GABAergic neuron subtypes. Chromatin immunoprecipitation and sequencing (ChIP-seq) analyses confirmed the binding of Dlx1 and Dlx2 near Gad1, Gad2, and Slc32a1 in forebrain GABAergic interneurons (Le et al., 2017; Pla et al., 2018; Lindtner et al., 2019). Dlx TF target enhancers are also found near Nrxn3 and Arx, which are important in GABAergic neuron maturation. Dlx1, Dlx2, and Dlx5 bind largely overlapping sites in the genome, and importantly, their binding can be associated with either an increase or a decrease in RE accessibility (Lindtner et al., 2019). Interestingly, Dlx TF-binding enhancers are highly enriched in the Ascl1-binding variant of E-box (Lindtner et al., 2019), consistent with a transcriptional relay. The availability of enhancers can also be modulated by Nkx2-2, Nkx2-1, and Lhx6 (Sandberg et al., 2016; Kim et al., 2021). It is not known where the other selector TFs bind.
It is not fully understood how the selector genes repress the alternative fate. In the mouse spinal cord, the neuromere-specific TFs Nkx2-2, Nkx6-1, and Olig2 negatively regulate each other’s expression and possibly some of their targets by directly binding to distinct REs (Kutejova et al., 2016). The DNA binding of Dlx TFs near genes that are involved in telencephalic neuroepithelium patterning (Otp, Gsh1, Ebf3, Gbx2, and Pax7) was associated with RE silencing (Lindtner et al., 2019). Perhaps the selector gene can negatively regulate the nuclear factors defining alternative fates, one transitional decision layer up. Alternatively, the activator TFs may simply be sequestered and RE silenced by an independent mechanism.
It is not fully clear if proneural genes and selector genes function independently or synergistically. Rather, both may be possible depending on the target gene. Examples are scarce, but there seems to be some differences between vertebrates and invertebrates in this case. In C. elegans, the proneural TFs are very often indispensable for the selector gene expression. For example, the expression of bHLH TFs Atonal (lin-32) and Achaete-Scute (hlh-14) precedes the expression of selector TFs in several cell types, including a POU-HD TF unc-86 in IL2 and URX neurons; DLX TF ceh-43 and SIX family TF ceh-32 in IL1 neurons, and an LHX TF lin-11 in AVJ neurons - and both the bHLH TF and the selector TF functions are required for proper differentiation (Masoudi et al., 2021). This is unlike the situation in mouse GABAergic neuron lineages. For instance, although Ascl1 can directly regulate Dlx1/2 expression in mouse telencephalon, expression of Dlx1/2 in telencephalon is delayed but not abolished in the absence of Ascl1 (Horton et al., 1999). The same is true for the expression of Gata2, Tal2, Six3, and Lhx1 in the diencephalic and midbrain GABAergic neurons in the Ascl1 mutant mouse (Peltopuro, Kala and Partanen, 2010; Virolainen et al., 2012). Possibly, the vertebrate animals feature more redundancy in bHLH TF function. Notably, Ascl1 and Pou3f2 function is synergistic in early neural gene activation (Fig. 1C).
Building and maintaining a competent state
Generic cofactor waves
As discussed, pioneer factor and selector gene potential is dependent on cellular competence. Aside the epigenetic state of chromatin, another important factor in defining and establishing competence is the availability of generic transcriptional regulators or cofactors.
Neuronal progenitor, precursor and neuron state descriptors can be defined as genes co-expressed in neuronal cells at different time points during and after cell-cycle exit. Some general characteristics of such temporal code have been described recently (Sagner et al., 2021) and are summarized in Table 1. In several regions of the neural tube, Onecut2(1/2/3) expression was found in neuronal precursors early, Pou2f2 and Zfhx3(3/4) in intermediate developmental stages, and Nfia, Nfib, and Neurod2/6 during the later stages of development (Sagner et al., 2021). Interestingly, CUT transcription factors are also expressed in all neuron types in C. elegans and control neuronal identity by regulating pan-neuronal gene expression in cooperation with the cell type-specific selector genes (Leyva-Díaz and Hobert, 2022). In mouse stem cell-derived neurons, the temporal code is subject to regulation by TGF-β signalling (Sagner et al., 2021). Nuclear Factor I (NFI) TFs are generic regulators of differentiation in several cell types (Chen et al., 2017). In neurons, Nfia and Nfib activate expression of Neurod2 via the same REs (Fig. 1C) (Sagner et al., 2021). NFI TFs may also function synergistically with other TFs to regulate early and late developmental genes. In cerebellar granule neurons, NFI proteins are recruited by Etv1 to the promoters of late differentiation genes, regulating synapse formation and maturation (Ding et al., 2016). Nfia mutant mice show severe defects in neurodevelopment and misregulation of genes associated with brain maturation (Wong et al., 2007). NeuroD genes are involved in regulation of neuronal migration. In E13.5 and E14.5 cortical subventricular zone neuronal precursors, Neurod1 is coexpressed with Tcf12 and Tcf4, and the Tcf12-Neurod1 interaction is important for cortical interneuron migration (Singh et al., 2022).
In the differentiation process, generic wave factors complement fate determinant function, may stabilize gene expression programs, and therefore can be seen as commitment factors. It is not known whether the wave is reversible or how it is tuned by external signalling. Mutations in the generic cofactor genes are often associated with brain malformations, neurodevelopmental delay linked to number of other developmental defects outside the nervous system (such as craniofacial development), or both (Table 1). This is expected given the generic cofactor and early developmental functions of the genes. The regulation of later-expressed cell type-specific genes in neurons may affect finer aspects of neurodevelopment, psychology, or behaviour. However, it is currently poorly understood how individual aspects of complex neuropsychiatric disorders or even features of normal behaviours are linked to the cell types in the brain.
Stepwise activation of terminal differentiation gene enhancers
As the chromatin state can be copied and maintained over cell divisions, the activity of temporally differentially expressed TFs can coregulate gene expression. For example, ASEL/ASER neurons, a morphologically symmetric neuron pair in C. elegans, arises from early separating progenitor lineages in development. At lineage separation, the TF tbx37/38 primes the lsy-6 promoter in ASEL progenitors for activation by che-1, a TF expressed later in the lineage, resulting in molecular and functional asymmetry (Charest et al., 2020). lsy-6 encodes a micro-RNA that targets an NKX homeobox TF cog-1 that, in turn, negatively regulates the expression of multiple genes, including the chemosensory receptor gcy-7 expressed in ASEL and the TF lim-6 that regulates the chemosensory receptor gcy-5 specific in ASER (Johnston and Hobert, 2003). che-1 and tbx37/38 are never coexpressed in ASER/ASEL lineages and priming of the lsy-6 promoter in ASEL is not affected by loss of any other TBX TF in C. elegans (Charest et al., 2020).
It would be interesting to further explore how early expressed TFs establish lineage-specific accessible states in gene loci or enhancers. One possible mechanism is the bivalent state via recruitment of histone modifiers. Early priming may explain how spatiotemporal code factors such as HOX, FOX, POU, LHX, and ETS TFs cooperate with the later-expressed selector TFs in vertebrates.
When continuous target expression is required, a feed-forward mechanism may be preferred. In developing neurons, once cell type-specific gene loci are activated, the maintenance of gene expression and accessible chromatin state is often relayed over from the initial selector genes to the same transcription factor family members. In the midbrain and diencephalic GABAergic neurons, the initial selector genes Gata2 and Tal2 are downregulated soon after the cell fate commitment, concomitant with the upregulation of Gata3 and Tal1, which could occupy the same enhancers in neuronal maturation stages (Kala et al., 2009; Achim et al., 2013). In forebrain GABAergic neurons, Dlx1/2 are expressed first, followed by Dlx5/6 expression (MacDonald et al., 2013). Relay to a different TF family member would require adaptation in RE sequences. Stage- or context-specific elements may cluster and form super-enhancers as seen in the retina of mice, where transcription of the Vsx2 gene requires activation of different stage-specific elements within a Vsx2 super-enhancer (Bian et al., 2022). Vsx2 is a selector-type TF that regulates both early and late-expressed genes in retinal bipolar neurons. Variation in RE sequences would allow maintenance of Vsx2 expression in subsequent stages of differentiation and its co-regulation with a temporally distinct set of target genes.
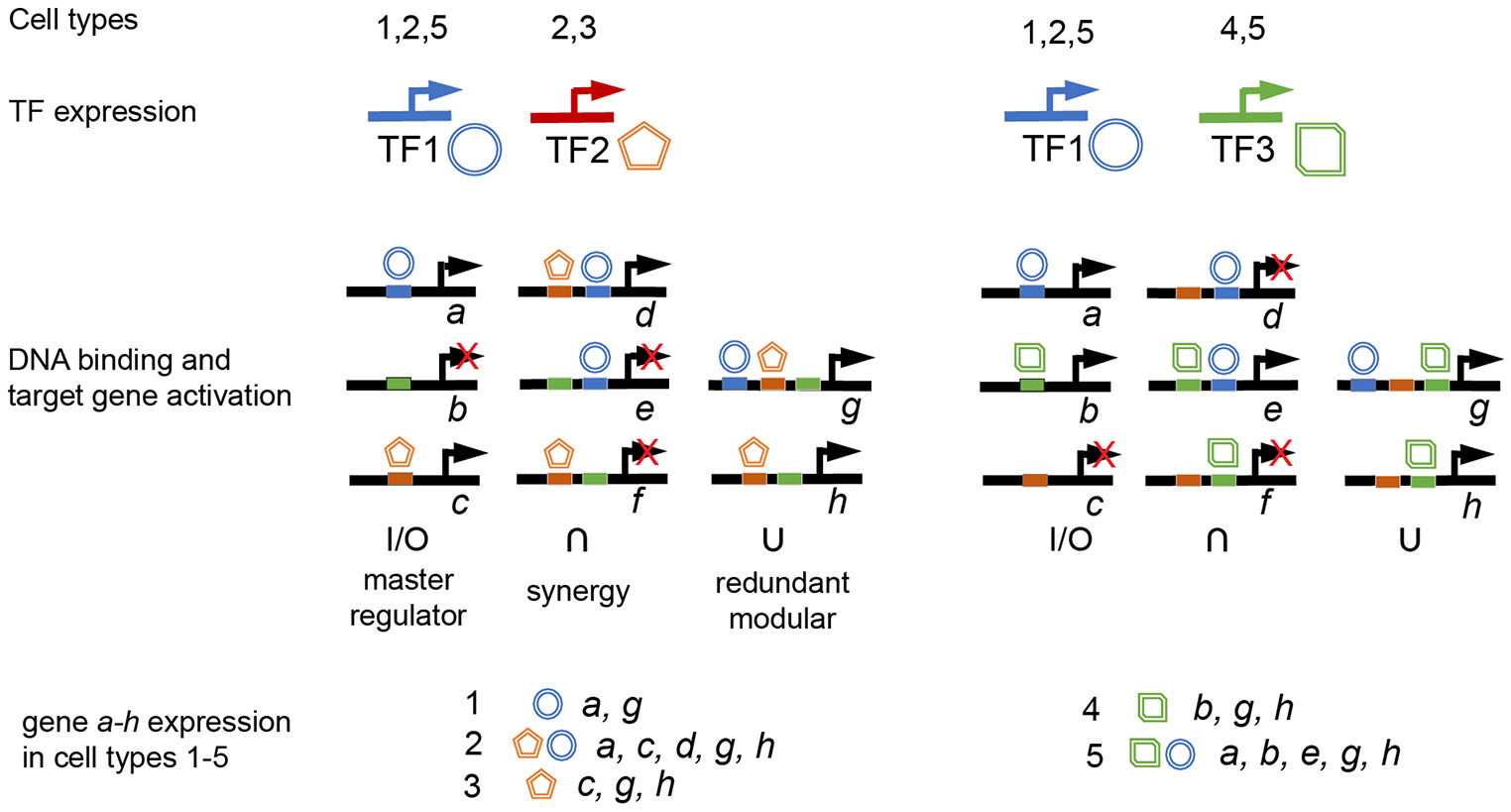
The compendium of TF-RE interactions across cell types and states is not yet fully explored. In addition to the number and sequence of REs and transcription factor availability, RE availability and the type of TF cooperation must be considered (Fig. 2). Single-cell multi-omics may have the potential to map the shift in active enhancers concomitant with the expression of transcriptional regulators and target gene expression, discovering regulatory interactions that can be tested in high-throughput assays such as Perturb-Seq (Peidli et al., 2024; Kim et al., 2024). The studies in the context of whole developing organisms or adult functions are yet to follow.
Fig. 2. Gene regulatory logic.
The combination of gene expression for 8 genes (a-h) in five cell types (1-5) expressing different combinations of transcription factors (TF1-TF3). All TFs are considered activating, and regulatory elements are considered available. Repressive interactions are not shown for simplicity. TF binding sites are indicated in matching color. Three enhancer types are shown. Master regulator: RE, where TF binding will lead to gene expression; Synergy: binding at all sites required for gene expression; Redundant modular: binding at any RE is sufficient for gene expression.
Remarks
Phenotypic convergence, similar to the concept of convergent evolution, refers to the phenomenon of similar cell types arising from molecularly distinct precursors. As such, phenotypic convergence has been discussed by studying the cell types derived from complex samples, such as whole embryos or the nervous system, using single-cell sequencing. However, perhaps the term should rather be used in reference to the cellular characteristics and not about the cell type as the unit.
Acknowledgements
I wish to thank the University of Helsinki Language Services for the language revision of the article.
Abbreviations
bHLH, basic helix-loop-helix ; C. elegans, Caenorhabditis elegans ; ChIP-seq, chromatin immunoprecipitation and sequencing ; CNS, central nervous system ; COE, Collier/Olf1/EBF family of transcription factors ; DHS, DNase I hypersensitivity sites ; Dll1, Delta-like 1 gene ; DNA, deoxyribonucleic acid ; E-box sequence, here, 5’-CANNTG-3’, the bHLH TF binding sequence in DNA ; ETS, E26 transformation-specific ; G2/M, Gap 2 to mitosis transition in cell cycle ; HD, homeodomain ; HD-CUT, CUT-homeobox protein ; HLH, helix-loop-helix protein domain ; HMG, high-mobility group ; iN, in vitro generated neurons ; iPSC, induced pluripotent stem cells ; kDa, kilodalton ; MEFs, mouse embryonic fibroblasts ; mESC, mouse embryonic stem cell ; NFI, nuclear factor I ; NPC, neural progenitor cells ; NR, nuclear receptors ; PNS, peripheral nervous system ; RE, regulatory element ; SOX, Sox family high-mobility group proteins ; TF, transcription factor ; ZF, zinc-finger domain ;References
Achim K., Peltopuro P., Lahti L., Tsai H.H., Zachariah A., Åstrand M., Salminen M., Rowitch D., Partanen J. (2013). The role of Tal2 and Tal1 in the differentiation of midbrain GABAergic neuron precursors. Biology Open 2: 990-997.
Achim K., Salminen M., Partanen J. (2014). Mechanisms regulating GABAergic neuron development. Cellular and Molecular Life Sciences 71: 1395-1415.
Al-Jawahiri R., Foroutan A., Kerkhof J., McConkey H., Levy M., Haghshenas S., Rooney K., Turner J., Shears D., Holder M., Lefroy H., Castle B., Reis L. M., Semina E. V., University of Washington Centre for Mendelian Genomics (UW-CMG) Lachlan K., Chandler K., Wright T., Clayton-Smith J., Hug F. P., McNeill A., (2022). SOX11 variants cause a neurodevelopmental disorder with infrequent ocular malformations and hypogonadotropic hypogonadism and with distinct DNA methylation profile. Genetics in medicine : official journal of the American College of Medical Genetics 24: 1261-1273.
Angelozzi M., Karvande A., Molin A. N., Ritter A. L., Leonard J. M. M., Savatt J. M., Douglass K., Myers S. M., Grippa M., Tolchin D., Zackai E., Donoghue S., Hurst A. C. E., Descartes M., Smith K., Velasco D., Schmanski A., Crunk A., Tokita M. J., de Lange I. M., van Gassen K., Robinson H., Guegan K., Suri M., Patel C., Bournez M., Faivre L., Tran-Mau-Them F., Baker J., Fabie N., Weaver K., Shillington A., Hopkin R. J., Barge-Schaapveld D. Q. C.M., Ruivenkamp C. A.L., Bökenkamp R., Vergano S., Seco Moro M. N., Díaz de Bustamante A., Misra V. K., Kennelly K., Rogers C., Friedman J., Wigby K. M., Lenberg J., Graziano C., Ahrens-Nicklas R. C., Lefebvre V. (2022). Consolidation of the clinical and genetic definition of a SOX4- related neurodevelopmental syndrome. Journal of Medical Genetics 59: 1058-1068.
Aydin B., Kakumanu A., Rossillo M., Moreno-Estellés M., Garipler G., Ringstad N., Flames N., Mahony S., Mazzoni E. O. (2019). Proneural factors Ascl1 and Neurog2 contribute to neuronal subtype identities by establishing distinct chromatin landscapes. Nature Neuroscience 22: 897-908.
Bian F., Daghsni M., Lu F., Liu S., Gross J. M., Aldiri I. (2022). Functional analysis of the Vsx2 super-enhancer uncovers distinct cis -regulatory circuits controlling Vsx2 expression during retinogenesis. Development 149: dev200642.
Bruce L., Peter B. (2022). Three children with different de novo BCL11A variants and diverse developmental phenotypes, but shared global motor discoordination and apraxic speech: Evidence for a functional gene network influencing the developing cerebellum and motor and auditory cortices. American Journal of Medical Genetics Part A 188: 3401-3415.
Castro D. S., Skowronska-Krawczyk D., Armant O., Donaldson I. J., Parras C., Hunt C., Critchley J. A., Nguyen L., Gossler A., Göttgens B., Matter J.M., Guillemot F. (2006). Proneural bHLH and Brn Proteins Coregulate a Neurogenic Program through Cooperative Binding to a Conserved DNA Motif. Developmental Cell 11: 831-844.
Chan M. M., Smith Z. D., Grosswendt S., Kretzmer H., Norman T. M., Adamson B., Jost M., Quinn J. J., Yang D., Jones M. G., Khodaverdian A., Yosef N., Meissner A., Weissman J. S. (2019). Molecular recording of mammalian embryogenesis. Nature 570: 77-82.
Charest J., Daniele T., Wang J., Bykov A., Mandlbauer A., Asparuhova M., Röhsner J., Gutiérrez-Pérez P., Cochella L. (2020). Combinatorial Action of Temporally Segregated Transcription Factors. Developmental Cell 55: 483-499.e7.
Chen C., Meng Q., Xia Y., Ding C., Wang L., Dai R., Cheng L., Gunaratne P., Gibbs R. A., Min S., Coarfa C., Reid J. G., Zhang C., Jiao C., Jiang Y., Giase G., Thomas A., Fitzgerald D., Brunetti T., Shieh A., Xia C., Wang Y., Wang Y., Badner J. A., Gershon E. S., White K. P., Liu C. (2018). The transcription factor POU3F2 regulates a gene coexpression network in brain tissue from patients with psychiatric disorders. Science Translational Medicine 10: eaat8178.
Chen K.S., Lim J. W.C., Richards L. J., Bunt J. (2017). The convergent roles of the nuclear factor I transcription factors in development and cancer. Cancer Letters 410: 124-138.
Chen X., Wyler S. C., Li L., Arnold A. G., Wan R., Jia L., Landy M. A., Lai H. C., Xu P., Liu C. (2020). Comparative Transcriptomic Analyses of Developing Melanocortin Neurons Reveal New Regulators for the Anorexigenic Neuron Identity. The Journal of Neuroscience 40: 3165-3177.
Colasante G., Lignani G., Rubio A., Medrihan L., Yekhlef L., Sessa A., Massimino L., Giannelli S. G., Sacchetti S., Caiazzo M., Leo D., Alexopoulou D., Dell’Anno M. T., Ciabatti E., Orlando M., Studer M., Dahl A., Gainetdinov R. R., Taverna S., Benfenati F., Broccoli V. (2015). Rapid Conversion of Fibroblasts into Functional Forebrain GABAergic Interneurons by Direct Genetic Reprogramming. Cell Stem Cell 17: 719-734.
Collins B. E., Neul J. L. (2022). Rett Syndrome and MECP2 Duplication Syndrome: Disorders of MeCP2 Dosage. Neuropsychiatric Disease and Treatment Volume 18: 2813-2835.
Coursimault J., Guerrot A. M., Morrow M. M., Schramm C., Zamora F. M., Shanmugham A., Liu S., Zou F., Bilan F., Le Guyader G., Bruel A. L., Denommé-Pichon A. S., Faivre L., Tran Mau-Them F., Tessarech M., Colin E., El Chehadeh S., Gérard B., Schaefer E., Cogne B., Lecoquierre F., (2022). MYT1L-associated neurodevelopmental disorder: description of 40 new cases and literature review of clinical and molecular aspects. Human genetics 141: 65-80.
Dias C., Estruch S. B., Graham S. A., McRae J., Sawiak S. J., Hurst J. A., Joss S. K., Holder S. E., Morton J. E.V., Turner C., Thevenon J., Mellul K., Sánchez-Andrade G., Ibarra-Soria X., Deriziotis P., Santos R. F., Lee S.C., Faivre L., Kleefstra T., Liu P., Hurles M. E., Fisher S. E., Logan D. W. (2016). BCL11A Haploinsufficiency Causes an Intellectual Disability Syndrome and Dysregulates Transcription. The American Journal of Human Genetics 99: 253-274.
Ding B., Cave J. W., Dobner P. R., Mullikin-Kilpatrick D., Bartzokis M., Zhu H., Chow C.W., Gronostajski R. M., Kilpatrick D. L. (2016). Reciprocal autoregulation by NFI occupancy and ETV1 promotes the developmental expression of dendrite-synapse genes in cerebellar granule neurons. Molecular Biology of the Cell 27: 1488-1499.
Doi D., Samata B., Katsukawa M., Kikuchi T., Morizane A., Ono Y., Sekiguchi K., Nakagawa M., Parmar M., Takahashi J. (2014). Isolation of Human Induced Pluripotent Stem Cell-Derived Dopaminergic Progenitors by Cell Sorting for Successful Transplantation. Stem Cell Reports 2: 337-350.
Fischer-Zirnsak B., Segebrecht L., Schubach M., Charles P., Alderman E., Brown K., Cadieux-Dion M., Cartwright T., Chen Y., Costin C., Fehr S., Fitzgerald K. M., Fleming E., Foss K., Ha T., Hildebrand G., Horn D., Liu S., Marco E. J., McDonald M., McWalter K., Race S., Rush E. T., Si Y., Saunders C., Slavotinek A., Stockler-Ipsiroglu S., Telegrafi A., Thiffault I., Torti E., Tsai A. C., Wang X., Zafar M., Keren B., Kornak U., Boerkoel C. F., Mirzaa G., Ehmke N. (2019). Haploinsufficiency of the Notch Ligand DLL1 Causes Variable Neurodevelopmental Disorders. The American Journal of Human Genetics 105: 631-639.
Fowler K. D., Funt J. M., Artyomov M. N., Zeskind B., Kolitz S. E., Towfic F. (2015). Leveraging existing data sets to generate new insights into Alzheimer’s disease biology in specific patient subsets. Scientific Reports 5: 14324.
Good K. V., Vincent J. B., Ausió J. (2021). MeCP2: The Genetic Driver of Rett Syndrome Epigenetics. Frontiers in Genetics 12: 620859.
Grosse M., Kuechler A., Dabir T., Spranger S., Beck-Wödl S., Bertrand M., Haack T. B., Grasemann C., Manka E., Depienne C., Kaiser F. J. (2023). Novel Variants of SOX4 in Patients with Intellectual Disability. International Journal of Molecular Sciences 24: 3519.
Habib A. M., Matsuyama A., Okorokov A. L., Santana-Varela S., Bras J. T., Aloisi A. M., Emery E. C., Bogdanov Y. D., Follenfant M., Gossage S. J., Gras M., Humphrey J., Kolesnikov A., Le Cann K., Li S., Minett M. S., Pereira V., Ponsolles C., Sikandar S., Torres J. M., Yamaoka K., Zhao J., Komine Y., Yamamori T., Maniatis N., Panov K. I., Houlden H., Ramirez J. D., Bennett D. L. H., Marsili L., Bachiocco V., Wood J. N., Cox J. J. (2018). A novel human pain insensitivity disorder caused by a point mutation in ZFHX2. Brain 141: 365-376.
Henke R. M., Savage T. K., Meredith D. M., Glasgow S. M., Hori K., Dumas J., MacDonald R. J., Johnson J. E. (2009). Neurog2 is a direct downstream target of the Ptf1a-Rbpj transcription complex in dorsal spinal cord. Development 136: 2945-2954.
Hoang C. Q., Hale M. A., Azevedo-Pouly A. C., Elsässer H. P., Deering T. G., Willet S. G., Pan F. C., Magnuson M. A., Wright C. V. E., Swift G. H., MacDonald R. J. (2016). Transcriptional Maintenance of Pancreatic Acinar Identity, Differentiation, and Homeostasis by PTF1A. Molecular and Cellular Biology 36: 3033-3047.
Hobert O., Kratsios P., (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Current opinion in neurobiology 56: 97-105.
Horton S., Meredith A., Richardson J. A., Johnson J. E. (1999). Correct Coordination of Neuronal Differentiation Events in Ventral Forebrain Requires the bHLH Factor MASH1. Molecular and Cellular Neuroscience 14: 355-369.
Huang H. S., Kubish G. M., Redmond T. M., Turner D. L., Thompson R. C., Murphy G. G., Uhler M. D. (2010). Direct transcriptional induction of Gadd45γ by Ascl1 during neuronal differentiation. Molecular and Cellular Neuroscience 44: 282-296.
Jacquemin P., Lannoy V. J., O'Sullivan J., Read A., Lemaigre F. P., Rousseau G. G. (2001). The Transcription Factor Onecut-2 Controls the Microphthalmia-Associated Transcription Factor Gene. Biochemical and Biophysical Research Communications 285: 1200-1205.
Jaura R., Yeh S.Y., Montanera K. N., Ialongo A., Anwar Z., Lu Y., Puwakdandawa K., Rhee H. S. (2022). Extended intergenic DNA contributes to neuron-specific expression of neighboring genes in the mammalian nervous system. Nature Communications 13: 2733.
Jin K., Xiang M. (2019). Transcription factor Ptf1a in development, diseases and reprogramming. Cellular and Molecular Life Sciences 76: 921-940.
Kala K., Haugas M., Lilleväli K., Guimera J., Wurst W., Salminen M., Partanen J. (2009). Gata2 is a tissue-specific post-mitotic selector gene for midbrain GABAergic neurons. Development 136: 253-262.
Kelberman D., de Castro S. C. P., Huang S., Crolla J. A., Palmer R., Gregory J. W., Taylor D., Cavallo L., Faienza M. F., Fischetto R., Achermann J. C., Martinez-Barbera J. P., Rizzoti K., Lovell-Badge R., Robinson I. C. A. F., Gerrelli D., Dattani M. T. (2008). SOX2 Plays a Critical Role in the Pituitary, Forebrain, and Eye during Human Embryonic Development. The Journal of Clinical Endocrinology & Metabolism 93: 1865-1873.
Kennedy-Williams P., Care H., Dalton L., Horton J., Kearney A., Rooney N., Hotton M., Pinckston M., Huggons E., Culshaw L., Kilcoyne S., Johnson D., Wilkie A. O.M., Wall S. (2021). Neurodevelopmental, Cognitive, and Psychosocial Outcomes for Individuals With Pathogenic Variants in the TCF12 Gene and Associated Craniosynostosis. Journal of Craniofacial Surgery 32: 1263-1268.
Kikuchi T., Morizane A., Doi D., Magotani H., Onoe H., Hayashi T., Mizuma H., Takara S., Takahashi R., Inoue H., Morita S., Yamamoto M., Okita K., Nakagawa M., Parmar M., Takahashi J. (2017). Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 548: 592-596.
Kim D. W., Liu K., Wang Z. Q., Zhang Y. S., Bathini A., Brown M. P., Lin S. H., Washington P. W., Sun C., Lindtner S., Lee B., Wang H., Shimogori T., Rubenstein J. L. R., Blackshaw S. (2021). Gene regulatory networks controlling differentiation, survival, and diversification of hypothalamic Lhx6-expressing GABAergic neurons. Communications Biology 4: 95.
Kim S. S., Truong B., Jagadeesh K., Dey K. K., Shen A. Z., Raychaudhuri S., Kellis M., Price A. L. (2024). Leveraging single-cell ATAC-seq and RNA-seq to identify disease-critical fetal and adult brain cell types. Nature Communications 15: 563.
Konstantinides N., Kapuralin K., Fadil C., Barboza L., Satija R., Desplan C. (2018). Phenotypic Convergence: Distinct Transcription Factors Regulate Common Terminal Features. Cell 174: 622-635.e13.
Kratsios P., Stolfi A., Levine M., Hobert O. (2012). Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nature Neuroscience 15: 205-214.
Krieger T. G., Moran C. M., Frangini A., Visser W. E., Schoenmakers E., Muntoni F., Clark C. A., Gadian D., Chong W. K., Kuczynski A., Dattani M., Lyons G., Efthymiadou A., Vargha-Khadem F., Simons B. D., Chatterjee K., Livesey F. J. (2019). Mutations in thyroid hormone receptor α1 cause premature neurogenesis and progenitor cell depletion in human cortical development. Proceedings of the National Academy of Sciences 116: 22754-22763.
Kutejova E., Sasai N., Shah A., Gouti M., Briscoe J. (2016). Neural Progenitors Adopt Specific Identities by Directly Repressing All Alternative Progenitor Transcriptional Programs. Developmental Cell 36: 639-653.
Labonne J. D. J., Shen Y., Kong I.K., Diamond M. P., Layman L. C., Kim H.G. (2016). Comparative deletion mapping at 1p31.3-p32.2 implies NFIA responsible for intellectual disability coupled with macrocephaly and the presence of several other genes for syndromic intellectual disability. Molecular Cytogenetics 9: 24.
Le T. N., Zhou Q.P., Cobos I., Zhang S., Zagozewski J., Japoni S., Vriend J., Parkinson T., Du G., Rubenstein J. L., Eisenstat D. D. (2017). GABAergic Interneuron Differentiation in the Basal Forebrain Is Mediated through Direct Regulation of Glutamic Acid Decarboxylase Isoforms by Dlx Homeobox Transcription Factors. The Journal of Neuroscience 37: 8816-8829.
Leyva-Díaz E., Hobert O. (2022). Robust regulatory architecture of pan-neuronal gene expression. Current Biology 32: 1715-1727.e8.
Limone F., Guerra San Juan I., Mitchell J. M., Smith J. L.M., Raghunathan K., Meyer D., Ghosh S. D., Couto A., Klim J. R., Joseph B. J., Gold J., Mello C. J., Nemesh J., Smith B. M., Verhage M., McCarroll S. A., Pietiläinen O., Nehme R., Eggan K. (2023). Efficient generation of lower induced motor neurons by coupling Ngn2 expression with developmental cues. Cell Reports 42: 111896.
Limone F., Klim J. R., Mordes D. A., (2022). Pluripotent stem cell strategies for rebuilding the human brain. Frontiers in aging neuroscience 14: 1017299.
Lin H.C., He Z., Ebert S., Schörnig M., Santel M., Nikolova M. T., Weigert A., Hevers W., Kasri N. N., Taverna E., Camp J. G., Treutlein B. (2021). NGN2 induces diverse neuron types from human pluripotency. Stem Cell Reports 16: 2118-2127.
Lindtner S., Catta-Preta R., Tian H., Su-Feher L., Price J. D., Dickel D. E., Greiner V., Silberberg S. N., McKinsey G. L., McManus M. T., Pennacchio L. A., Visel A., Nord A. S., Rubenstein J. L.R. (2019). Genomic Resolution of DLX-Orchestrated Transcriptional Circuits Driving Development of Forebrain GABAergic Neurons. Cell Reports 28: 2048-2063.e8.
Liu C., Maejima T., Wyler S. C., Casadesus G., Herlitze S., Deneris E. S. (2010). Pet-1 is required across different stages of life to regulate serotonergic function. Nature Neuroscience 13: 1190-1198.
Liu X., Li F., Stubblefield E. A., Blanchard B., Richards T. L., Larson G. A., He Y., Huang Q., Tan A. C., Zhang D., Benke T. A., Sladek J. R., Zahniser N. R., Li C. Y., (2012). Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell research 22: 321-332.
MacDonald R. B., Pollack J. N., Debiais-Thibaud M., Heude E., Coffin Talbot J., Ekker M. (2013). The ascl1a and dlx genes have a regulatory role in the development of GABAergic interneurons in the zebrafish diencephalon. Developmental Biology 381: 276-285.
Macrae T. A., Fothergill-Robinson J., Ramalho-Santos M. (2023). Regulation, functions and transmission of bivalent chromatin during mammalian development. Nature Reviews Molecular Cell Biology 24: 6-26.
Mall M., Kareta M. S., Chanda S., Ahlenius H., Perotti N., Zhou B., Grieder S. D., Ge X., Drake S., Euong Ang C., Walker B. M., Vierbuchen T., Fuentes D. R., Brennecke P., Nitta K. R., Jolma A., Steinmetz L. M., Taipale J., Südhof T. C., Wernig M., (2017). Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 544: 245-249.
Masoudi N., Yemini E., Schnabel R., Hobert O. (2021). Piecemeal regulation of convergent neuronal lineages by bHLH transcription factors in Caenorhabditis elegans. Development 148: dev199224.
Masserdotti G., Gillotin S., Sutor B., Drechsel D., Irmler M., Jørgensen H. F., Sass S., Theis F. J., Beckers J., Berninger B., Guillemot F., Götz M. (2015). Transcriptional Mechanisms of Proneural Factors and REST in Regulating Neuronal Reprogramming of Astrocytes. Cell Stem Cell 17: 74-88.
Melis D., Carvalho D., Barbaro‐Dieber T., Espay A. J., Gambello M. J., Gener B., Gerkes E., Hitzert M. M., Hove H. B., Jansen S., Jira P. E., Lachlan K., Menke L. A., Narayanan V., Ortiz D., Overwater E., Posmyk R., Ramsey K., Rossi A., Sandoval R. L., Stumpel C., Stuurman K. E., Cordeddu V., Turnpenny P., Strisciuglio P., Tartaglia M., Unger S., Waters T., Turnbull C., Hennekam R. C. (2020). Primrose syndrome: Characterization of the phenotype in 42 patients. Clinical Genetics 97: 890-901.
Mora-Martinez C. (2021). Expression pattern determines regulatory logic. PLOS ONE 16: e0244864.
Negishi Y., Miya F., Hattori A., Mizuno K., Hori I., Ando N., Okamoto N., Kato M., Tsunoda T., Yamasaki M., Kanemura Y., Kosaki K., Saitoh S. (2015). Truncating mutation in NFIA causes brain malformation and urinary tract defects. Human Genome Variation 2: 15007.
Norrie J. L., Lupo M. S., Xu B., Al Diri I., Valentine M., Putnam D., Griffiths L., Zhang J., Johnson D., Easton J., Shao Y., Honnell V., Frase S., Miller S., Stewart V., Zhou X., Chen X., Dyer M. A. (2019). Nucleome Dynamics during Retinal Development. Neuron 104: 512-528.e11.
Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C., Wernig M. (2011). Induction of human neuronal cells by defined transcription factors. Nature 476: 220-223.
Papes F., Camargo A. P., de Souza J. S., Carvalho V. M. A., Szeto R. A., LaMontagne E., Teixeira J. R., Avansini S. H., Sánchez-Sánchez S. M., Nakahara T. S., Santo C. N., Wu W., Yao H., Araújo B. M. P., Velho P. E. N. F., Haddad G. G., Muotri A. R. (2022). Transcription Factor 4 loss-of-function is associated with deficits in progenitor proliferation and cortical neuron content. Nature Communications 13: 2387.
Peidli S., Green T. D., Shen C., Gross T., Min J., Garda S., Yuan B., Schumacher L. J., Taylor-King J. P., Marks D. S., Luna A., Blüthgen N., Sander C. (2024). scPerturb: harmonized single-cell perturbation data. Nature Methods 21: 531-540.
Peltopuro P., Kala K., Partanen J. (2010). Distinct requirements for Ascl1 in subpopulations of midbrain GABAergic neurons. Developmental Biology 343: 63-70.
Pla R., Stanco A., Howard M.K. A., Rubin A. N., Vogt D., Mortimer N., Cobos I., Potter G. B., Lindtner S., Price J. D., Nord A. S., Visel A., Schreiner C. E., Baraban S. C., Rowitch D. H., Rubenstein J. L. R. (2018). Dlx1 and Dlx2 Promote Interneuron GABA Synthesis, Synaptogenesis, and Dendritogenesis. Cerebral Cortex 28: 3797-3815.
Popp B., Bienvenu T., Giurgea I., Metreau J., Kraus C., Reis A., Fischer J., Bralo M. P., Tenorio‐Castaño J., Lapunzina P., Almoguera B., Lopez‐Grondona F., Sticht H., Zweier C. (2022). The recurrent TCF4 missense variant p.( Arg389Cys ) causes a neurodevelopmental disorder overlapping with but not typical for Pitt‐Hopkins syndrome. Clinical Genetics 102: 517-523.
Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., Wysocka J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279-283.
Rada-Iglesias A., Bajpai R., Prescott S., Brugmann S. A., Swigut T., Wysocka J. (2012). Epigenomic Annotation of Enhancers Predicts Transcriptional Regulators of Human Neural Crest. Cell Stem Cell 11: 633-648.
Raposo A. A.S.F., Vasconcelos F. F., Drechsel D., Marie C., Johnston C., Dolle D., Bithell A., Gillotin S., van den Berg D. L.C., Ettwiller L., Flicek P., Crawford G. E., Parras C. M., Berninger B., Buckley N. J., Guillemot F., Castro D. S. (2015). Ascl1 Coordinately Regulates Gene Expression and the Chromatin Landscape during Neurogenesis. Cell Reports 10: 1544-1556.
Rhee H. S., Closser M., Guo Y., Bashkirova E. V., Tan G. C., Gifford D. K., Wichterle H. (2016). Expression of Terminal Effector Genes in Mammalian Neurons Is Maintained by a Dynamic Relay of Transient Enhancers. Neuron 92: 1252-1265.
Sagner A., Zhang I., Watson T., Lazaro J., Melchionda M., Briscoe J. (2021). A shared transcriptional code orchestrates temporal patterning of the central nervous system. PLOS Biology 19: e3001450.
Sandberg M., Flandin P., Silberberg S., Su-Feher L., Price J. D., Hu J. S., Kim C., Visel A., Nord A. S., Rubenstein J. L.R. (2016). Transcriptional Networks Controlled by NKX2-1 in the Development of Forebrain GABAergic Neurons. Neuron 91: 1260-1275.
Satoh J., Yamamoto Y., Asahina N., Kitano S., Kino Y. (2014). RNA-Seq Data Mining: Downregulation of NeuroD6 Serves as a Possible Biomarker for Alzheimer’s Disease Brains. Disease Markers 2014: 1-10.
Schanze I., Bunt J., Lim J. W.C., Schanze D., Dean R. J., Alders M., Blanchet P., Attié-Bitach T., Berland S., Boogert S., Boppudi S., Bridges C. J., Cho M. T., Dobyns W. B., Donnai D., Douglas J., Earl D. L., Edwards T. J., Faivre L., Fregeau B., Genevieve D., Gérard M., Gatinois V., Holder-Espinasse M., Huth S. F., Izumi K., Kerr B., Lacaze E., Lakeman P., Mahida S., Mirzaa G. M., Morgan S. M., Nowak C., Peeters H., Petit F., Pilz D. T., Puechberty J., Reinstein E., Rivière J.B., Santani A. B., Schneider A., Sherr E. H., Smith-Hicks C., Wieland I., Zackai E., Zhao X., Gronostajski R. M., Zenker M., Richards L. J. (2018). NFIB Haploinsufficiency Is Associated with Intellectual Disability and Macrocephaly. The American Journal of Human Genetics 103: 752-768.
Schönauer R., Jin W., Findeisen C., Valenzuela I., Devlin L. A., Murrell J., Bedoukian E. C., Pöschla L., Hantmann E., Riedhammer K. M., Hoefele J., Platzer K., Biemann R., Campeau P. M., Münch J., Heyne H., Hoffmann A., Ghosh A., Sun W., Dong H., Noé F., Wolfrum C., Woods E., Parker M. J., Neatu R., Le Guyader G., Bruel A.L., Perrin L., Spiewak H., Missotte I., Fourgeaud M., Michaud V., Lacombe D., Paolucci S. A., Buchan J. G., Glissmeyer M., Popp B., Blüher M., Sayer J. A., Halbritter J. (2023). Monoallelic intragenic POU3F2 variants lead to neurodevelopmental delay and hyperphagic obesity, confirming the gene’s candidacy in 6q16.1 deletions. The American Journal of Human Genetics 110: 998-1007.
Sega A. G., Mis E. K., Lindstrom K., Mercimek-Andrews S., Ji W., Cho M. T., Juusola J., Konstantino M., Jeffries L., Khokha M. K., Lakhani S. A. (2019). De novo pathogenic variants in neuronal differentiation factor 2 (NEUROD2) cause a form of early infantile epileptic encephalopathy. Journal of Medical Genetics 56: 113-122.
Seo S., Lim J.W., Yellajoshyula D., Chang L.W., Kroll K. L. (2007). Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. The EMBO Journal 26: 5093-5108.
Serrano-Saiz E., Poole R. J., Felton T., Zhang F., De La Cruz E. D., Hobert O. (2013). Modular Control of Glutamatergic Neuronal Identity in C. elegans by Distinct Homeodomain Proteins. Cell 155: 659-673.
Sharma V. P., Fenwick A. L., Brockop M. S., McGowan S. J., Goos J. A. C., Hoogeboom A. J. M., Brady A. F., Jeelani N. O., Lynch S. A., Mulliken J. B., Murray D. J., Phipps J. M., Sweeney E., Tomkins S. E., Wilson L. C., Bennett S., Cornall R. J., Broxholme J., Kanapin A., Johnson D., Wall S. A., van der Spek P. J., Mathijssen I. M. J., Maxson R. E., Twigg S. R. F., Wilkie A. O. M. (2013). Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nature Genetics 45: 304-307.
Singh A., Mahesh A., Noack F., Cardoso de Toledo B., Calegari F., Tiwari V. K. (2022). Tcf12 and NeuroD1 cooperatively drive neuronal migration during cortical development. Development 149: dev200250.
Song Y., Soto J., Chen B., Hoffman T., Zhao W., Zhu N., Peng Q., Liu L., Ly C., Wong P. K., Wang Y., Rowat A. C., Kurdistani S. K., Li S. (2022). Transient nuclear deformation primes epigenetic state and promotes cell reprogramming. Nature Materials 21: 1191-1199.
Soufi A., Donahue G., Zaret K. S. (2012). Facilitators and Impediments of the Pluripotency Reprogramming Factors' Initial Engagement with the Genome. Cell 151: 994-1004.
Spellmann I., Riedel M., Städtler J., Zill P., Obermeier M., Cerovecki A., Dehning S., Schennach R., Epple M., Opgen-Rhein M., Müller N., Bondy B., Möller H.J., Musil R. (2017). Associations of NEUROD2 polymorphisms and change of cognitive dysfunctions in schizophrenia and schizoaffective disorder after eight weeks of antipsychotic treatment. Cognitive Neuropsychiatry 22: 280-297.
Stefanakis N., Carrera I., Hobert O. (2015). Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron 87: 733-750.
Sun A. X., Yuan Q., Tan S., Xiao Y., Wang D., Khoo A. T. T., Sani L., Tran H.D., Kim P., Chiew Y. S., Lee K. J., Yen Y.C., Ng H. H., Lim B., Je H. S. (2016). Direct Induction and Functional Maturation of Forebrain GABAergic Neurons from Human Pluripotent Stem Cells. Cell Reports 16: 1942-1953.
Tamura R. E., de Vasconcellos J. F., Sarkar D., Libermann T. A., Fisher P. B., Zerbini L. F. (2012). GADD45 Proteins: Central Players in Tumorigenesis. Current Molecular Medicine 12: 634-651.
Tanaka S., Kodama T., Nonaka T., Toyoda H., Arai M., Fukazawa M., Honda Y., Honda M., Mignot E. (2010). Transcriptional regulation of the hypocretin/orexin gene by NR6A1. Biochemical and Biophysical Research Communications 403: 178-183.
Thoma E. C., Wischmeyer E., Offen N., Maurus K., Sirén A.L., Schartl M., Wagner T. U. (2012). Ectopic Expression of Neurogenin 2 Alone is Sufficient to Induce Differentiation of Embryonic Stem Cells into Mature Neurons. PLoS ONE 7: e38651.
Tolve M., Ulusoy A., Patikas N., Islam K. U. S., Bodea G. O., Öztürk E., Broske B., Mentani A., Wagener A., van Loo K. M.J., Britsch S., Liu P., Khaled W. T., Metzakopian E., Baader S. L., Di Monte D. A., Blaess S. (2021). The transcription factor BCL11A defines distinct subsets of midbrain dopaminergic neurons. Cell Reports 36: 109697.
Tsunemoto R., Lee S., Szűcs A., Chubukov P., Sokolova I., Blanchard J. W., Eade K. T., Bruggemann J., Wu C., Torkamani A., Sanna P. P., Baldwin K. K. (2018). Diverse reprogramming codes for neuronal identity. Nature 557: 375-380.
Uzman C. Y., Gürsoy S., Hazan F. (2023). A rare cause of intellectual disability: Novel mutations of NFIX gene in two patients with clinical features of Marshall–Smith syndrome and Malan syndrome. International Journal of Developmental Neuroscience 83: 479-485.
Virolainen S.M., Achim K., Peltopuro P., Salminen M., Partanen J. (2012). Transcriptional regulatory mechanisms underlying the GABAergic neuron fate in different diencephalic prosomeres. Development 139: 3795-3805.
Wagner D. E., Weinreb C., Collins Z. M., Briggs J. A., Megason S. G., Klein A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360: 981-987.
Wang Q., Wu J., Yang J., Huang S., Yuan Y., Dai P. (2023). Two SOX11 variants cause Coffin–Siris syndrome with a new feature of sensorineural hearing loss. American Journal of Medical Genetics Part A 191: 183-189.
Wapinski O. L., Lee Q. Y., Chen A. C., Li R., Corces M. R., Ang C. E., Treutlein B., Xiang C., Baubet V., Suchy F. P., Sankar V., Sim S., Quake S. R., Dahmane N., Wernig M., Chang H. Y. (2017). Rapid Chromatin Switch in the Direct Reprogramming of Fibroblasts to Neurons. Cell Reports 20: 3236-3247.
Wapinski O. L., Vierbuchen T., Qu K., Lee Q. Y., Chanda S., Fuentes D. R., Giresi P. G., Ng Y. H., Marro S., Neff N. F., Drechsel D., Martynoga B., Castro D. S., Webb A. E., Südhof T. C., Brunet A., Guillemot F., Chang H. Y., Wernig M. (2013). Hierarchical Mechanisms for Direct Reprogramming of Fibroblasts to Neurons. Cell 155: 621-635.
Wong Y., Schulze C., Streichert T., Gronostajski R. M., Schachner M., Tilling T. (2007). Gene expression analysis of nuclear factor I-A deficient mice indicates delayed brain maturation. Genome Biology 8: R72.
Woodworth M. B., Greig L. C., Liu K. X., Ippolito G. C., Tucker H. O., Macklis J. D. (2016). Ctip1 Regulates the Balance between Specification of Distinct Projection Neuron Subtypes in Deep Cortical Layers. Cell Reports 15: 999-1012.
Wyler S. C., Spencer W. C., Green N. H., Rood B. D., Crawford L.T., Craige C., Gresch P., McMahon D. G., Beck S. G., Deneris E. (2016). Pet-1 Switches Transcriptional Targets Postnatally to Regulate Maturation of Serotonin Neuron Excitability. The Journal of Neuroscience 36: 1758-1774.
Yu Y., Li X., Jiao R., Lu Y., Jiang X., Li X. (2023). H3K27me3-H3K4me1 transition at bivalent promoters instructs lineage specification in development. Cell & Bioscience 13: 66.
Zenteno J. C., Perez‐Cano H. J., Aguinaga M. (2006). Anophthalmia‐esophageal atresia syndrome caused by an SOX2 gene deletion in monozygotic twin brothers with markedly discordant phenotypes. American Journal of Medical Genetics Part A 140A: 1899-1903.
Zhu F., Farnung L., Kaasinen E., Sahu B., Yin Y., Wei B., Dodonova S. O., Nitta K. R., Morgunova E., Taipale M., Cramer P., Taipale J. (2018). The interaction landscape between transcription factors and the nucleosome. Nature 562: 76-81.