Int. J. Dev. Biol. 66: 383 - 389 (2022)
The involvement of hormone-sensitive lipase in all-trans retinoic acid induced cleft palate
Original Article | Published: 20 January 2023
Abstract
Abnormally high concentrations of all-trans retinoic acid (atRA) induce cleft palate, which is accompanied by abnormal migration and proliferation of mouse embryonic palatal mesenchyme (MEPM) cells. Hormone-sensitive lipase (HSL) is involved in many embryonic development processes. The current study was designed to elucidate the mechanism of HSL in cleft palate induced by atRA. To establish a cleft palate model in Kunming mice, pregnant mice were administered atRA (70 mg/kg) by gavage at embryonic Day 10.5 (E10.5). Embryonic palates were obtained through the dissection of pregnant mice at E15.5. Hematoxylin and eosin (H&E) staining was used to evaluate growth changes in the palatal shelves. The levels of HSL in MEPM cells were detected by immunohistochemistry, quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) and western blotting. RNAi was applied to construct vectors expressing HSL small interference RNAs (siRNAs). The vectors were transfected into MEPM cells. Cell proliferation and migration were evaluated by the cell counting kit-8 (CCK-8) assay and wound healing assay, respectively. The palatal shelves in the atRA group had separated at E15.5 without fusing. In MEPM cells, the expression of HSL was reversed after atRA treatment, which caused cleft palate in vivo. In the atRA group, the proliferation of HSL siRNA-transfected cells was remarkably promoted, and the migration rate significantly increased in the HSL siRNA-transfected MEPM cells. These results suggested that HSL may be involved in cleft palate induced by atRA and that atRA enhances HSL levels to inhibit embryonic palate growth.
Keywords
Cleft palate, atRA, HSL, MEPM
Introduction
Cleft lip and palate are among the most common congenital diseases in humans, resulting from environmental or genetic factors of facial development in utero (Alois et al., 2020). Facial morphogenesis occurs at the early stages of embryonic development. In this process, the paired maxillary and mandibular and the medial and lateral nasal prominences constitute the first partly separate facial primordia (Worley et al., 2018). Palatogenesis starts at week 4 in human embryonic development and is not completed until approximately week 12 (Mossey et al., 2009). The palate consists of three elements: the primary palate, which derives from the frontonasal process, and the lateral protrusion of the maxillary processes will form a pair of secondary palates; the development of the secondary palate is a multistep process that consists of palatal shelf growth, the elevation of palatal shelves, the fusion between the paired palatal shelves, and the disappearance of the medial epithelial seam; and the closed palate separates the oropharynx from the nasopharynx, the palatal shelves grow into the midline, and palatal shelves are fully formed (Abramyan et al., 2015; Nohara et al., 2022). Mice have widely been used for palatal development studies. Palate formation and related molecular mechanisms in mice are similar to those in humans (Suzuki et al., 2018). In mice, the secondary palate development begins at embryonic Day 11.5 (E11.5), and at E14.5 and 15.5, the two palatal shelves each meet and fuse in the middle of the oral cavity. Palate formation drastically changes, between E14.5 and E15.5, from the cell proliferation phase to differentiation; not only morphology but also the gene expression is altered according to cellular events (Bush et al., 2012). Any imbalance in embryonic palatal mesenchyme cell proliferation and apoptosis can result in cleft palate formation (Gao et al., 2016).
Lipids, due to their multiple biological roles in cell biology, physiology, and pathology, are crucial for energy storage and are also involved in cell signaling as well as in gene expression during embryonic development. Several human diseases have been found to be associated with abnormal lipid levels (Knittel et al., 2022). Hormone-sensitive lipase (HSL) is a crucial enzyme in regulating lipid metabolism. According to its physiological properties, HSL is an intracellular neutral lipase that is highly expressed in adipose tissue and can catalyze the hydrolysis of triacylglycerol and diacylglycerol (Sztrolovics et al., 1997). According to some studies, the catalytic function of HSL is essential for fertility in male mice (Wang et al., 2014), and 2,4-dienoyl-CoA reductase (DECR1) directly activates HSL to promote lipolysis in cervical cancer cells to support cervical cancer cell growth (Zhou et al., 2022). A previous study also showed the relationship between the incidence of cleft palate in fetuses and the change in endogenous small molecular metabolites. Additional research on the expression of related genes in lipid metabolism will likely determine the pathogenetic mechanisms of craniofacial malformation (Zhou et al., 2011).
Retinoic acid (RA) is a synthetic retinoid used to treat psoriasis. All-trans retinoic acid (atRA) is an oxidant of RA and supports normal pattern formation during embryogenesis (Al Tanoury et al., 2013). In both experimental animals and humans, abnormally high concentrations of atRA induce fetal malformations, including cleft palate (Ackermans et al., 2011). Previous in vivo studies have demonstrated that atRA induces growth retardation of the palatal shelves (Wang et al., 2017). In the present study, we created cleft palate models in Kunming mice using abnormally high concentrations of atRA to test whether the expression of HSL differs from that of the control group. In addition, we investigated whether HSL affects the migration and proliferation of MEPM cells in the atRA group.
Results
Histology of embryonic palate shelves
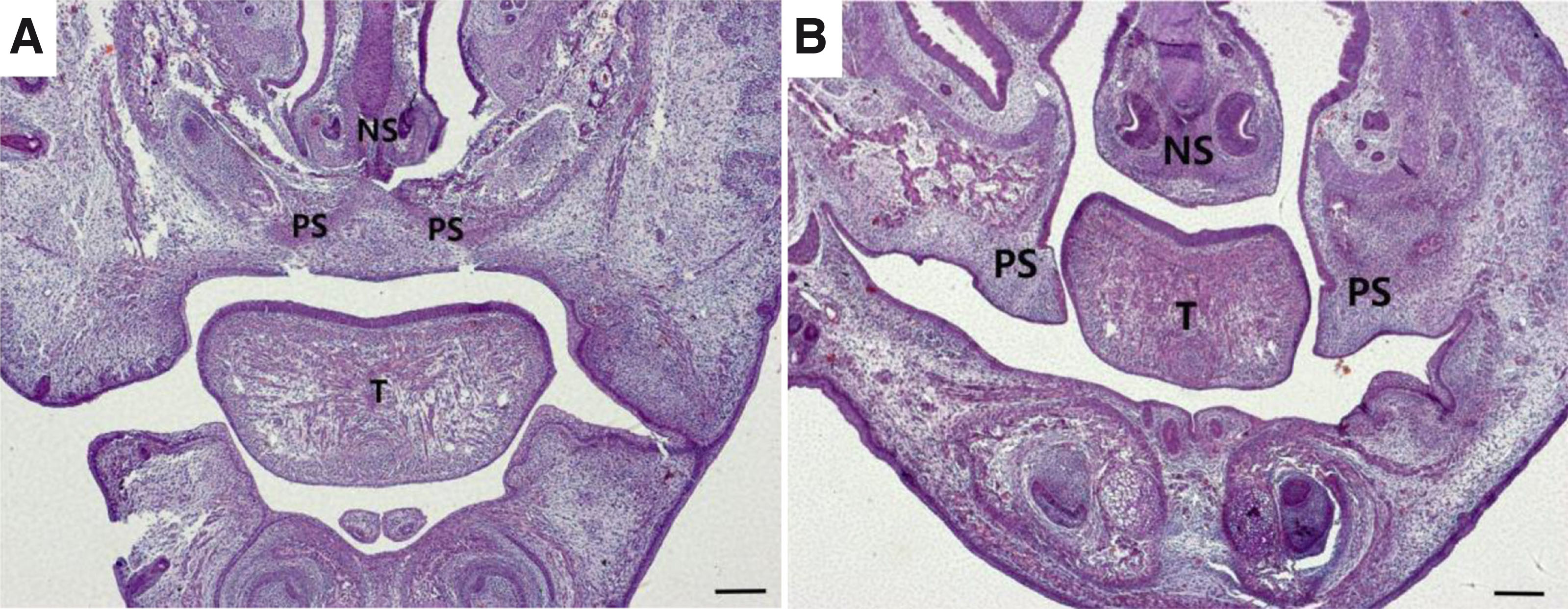
Embryonic palate shelf tissue was collected from pregnant mice. The palate shelf tissue and histological sections of the control group showed that the palatal shelf fused and formed a complete palate. The midline epithelial seam (MES) had disappeared in the mid and anterior regions at E15.5. The atRA-treated group showed that the palatal shelf was separated without fusion, and the palatal processes on both sides were short. A broad cleft phenotype was present in the atRA group (Fig. 1).
Fig. 1. Hematoxylin and eosin (H&E) staining of palate shelf tissues at E15.5 between control vs. atRA-treated mice
(A) The palatal shelf had been fused and eventually formed a complete palate. The midline epithelial seam (MES) disappeared in the mid anterior region; (B) Unfused, separated palatal shelf from an embryo of an atRA-treated mouse, the palatal processes on both sides were short. A broad cleft phenotype was eventually presented. (A, B) H&E staining images (scale bar, 400 µm). PS, palatal shelf; T, tongue; NS, nasal septum; H&E, hematoxylin, and eosin.
HSL significantly increases the MEPM of cleft palate
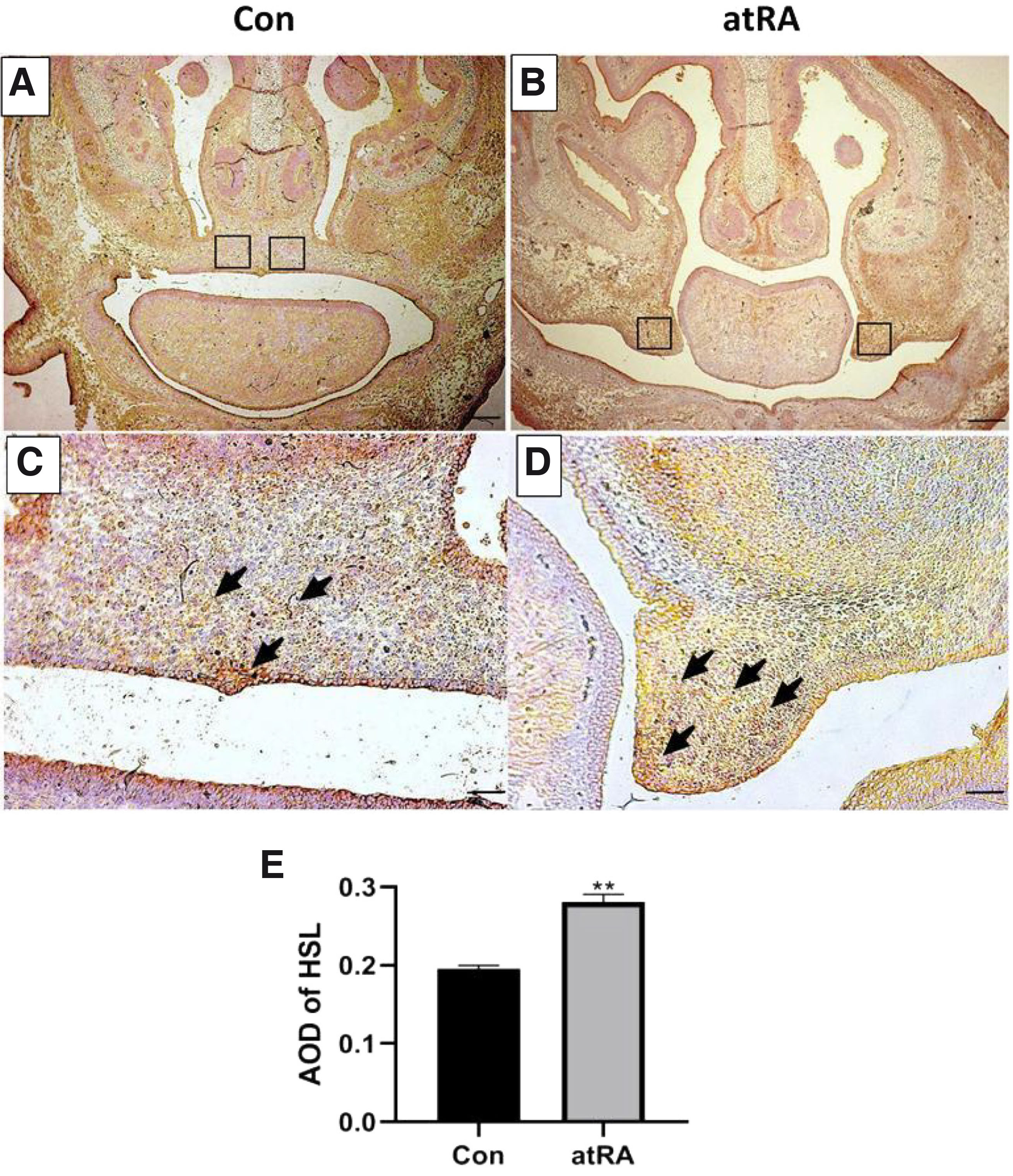
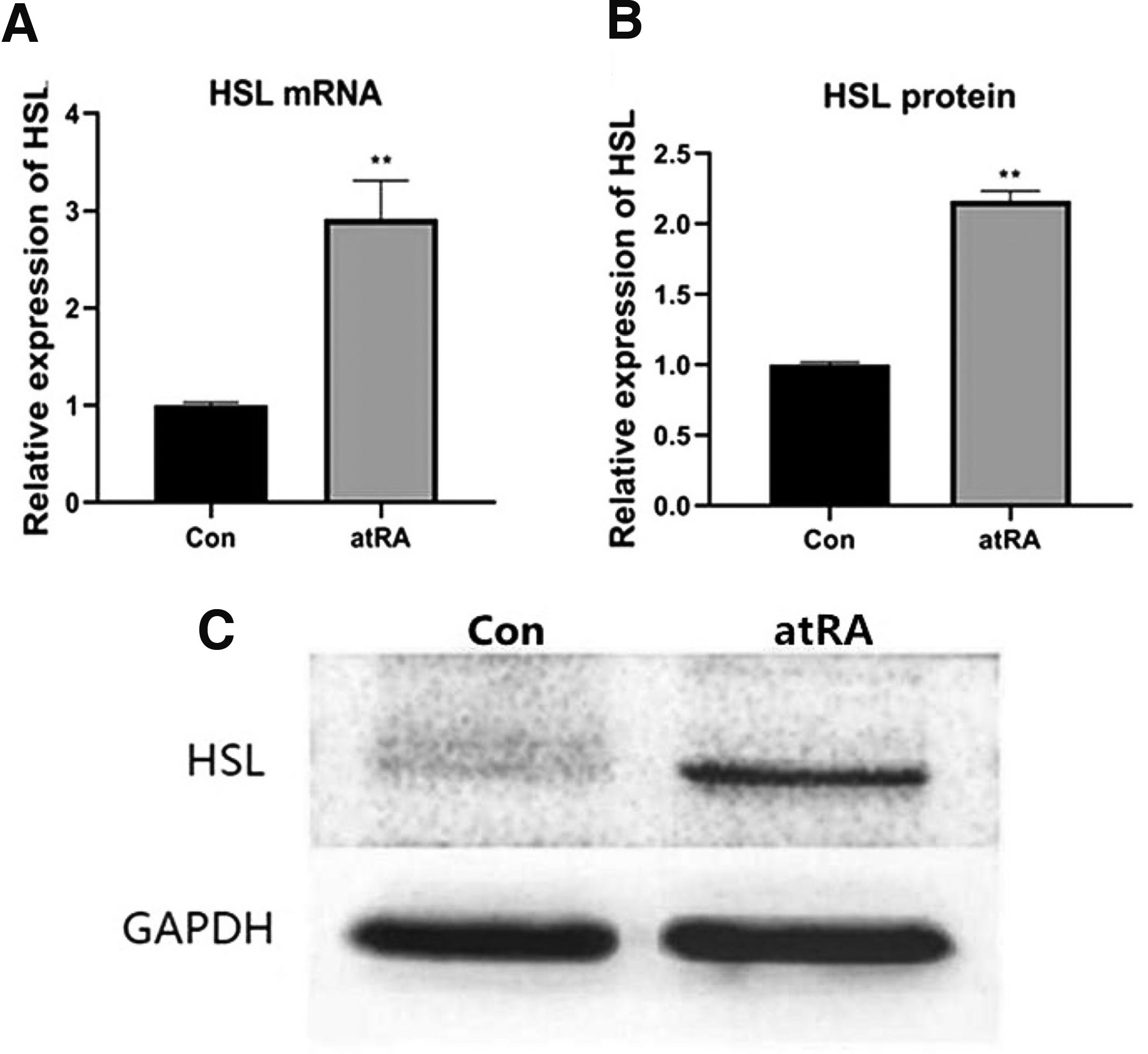
At E15.5, the palate shelves had fused in the control group (Fig. 2A). In contrast, the palatal shelves appeared abnormally small and failed to undergo elevation and fusion, finally developing into cleft palate in the atRA-treated group at E15.5 (Fig. 2B). There were more HSL-positive cells in the atRA group than in the control group (Fig. 2 C,D). The expression of HSL protein was upregulated in the MEPM cells in the atRA treatment group compared with the control group (Fig. 2E; **P<0.01). The qRT-PCR results indicated that the mRNA level of HSL was upregulated at E15.5 in the atRA group compared with the control group (Fig. 3A; **P<0.01). The western blot results showed that the protein levels of HSL were significantly increased in the atRA-exposed MEPM cells compared with the control MEPM cells at E15.5 (Fig. 3 B,C; **P<0.01).
Fig. 2. Immunohistochemistry assay demonstrating HSL expression in the EPM cells.
HSL was significantly increased at E15.5 in the MEPM cells in the atRA group compared to the Con group. (A, B) Immunohistochemistry results (scale bar, 400 µm). (C, D) Immunohistochemistry results (scale bar, 100 µm). (C, arrowhead)In the control group, HSL-positive cells were found in the palatal fusion area, (D, arrowhead). In the atRA group, the surface of the unilateral palatal process epithelium showed obvious yellowish-brown and scattered brown yellow areas were observed in the palatal process mesenchyme. (E) HSL AOD quantification. **P<0.01. HSL, hormone-sensitive lipase; AOD, average optical density; MEPM, mouse embryonic palatal mesenchyme; E, embryonic day; atRA, all-trans retinoic acid; Con, control.
Fig. 3. The expression of HSL was upregulated in the atRA group.
(A) When compared with the Con group, mRNA expression of HSL was increased in the atRA group at E15.5. (B, C) The protein expression of HSL was higher in the atRA group than in the Con group. **P<0.01. HSL, hormone-sensitive lipase; MEPM, mouse embryonic palatal mesenchyme; atRA, all-trans retinoic acid; E, embryonic day; Con, control.
Silencing of the HSL gene by RNAi
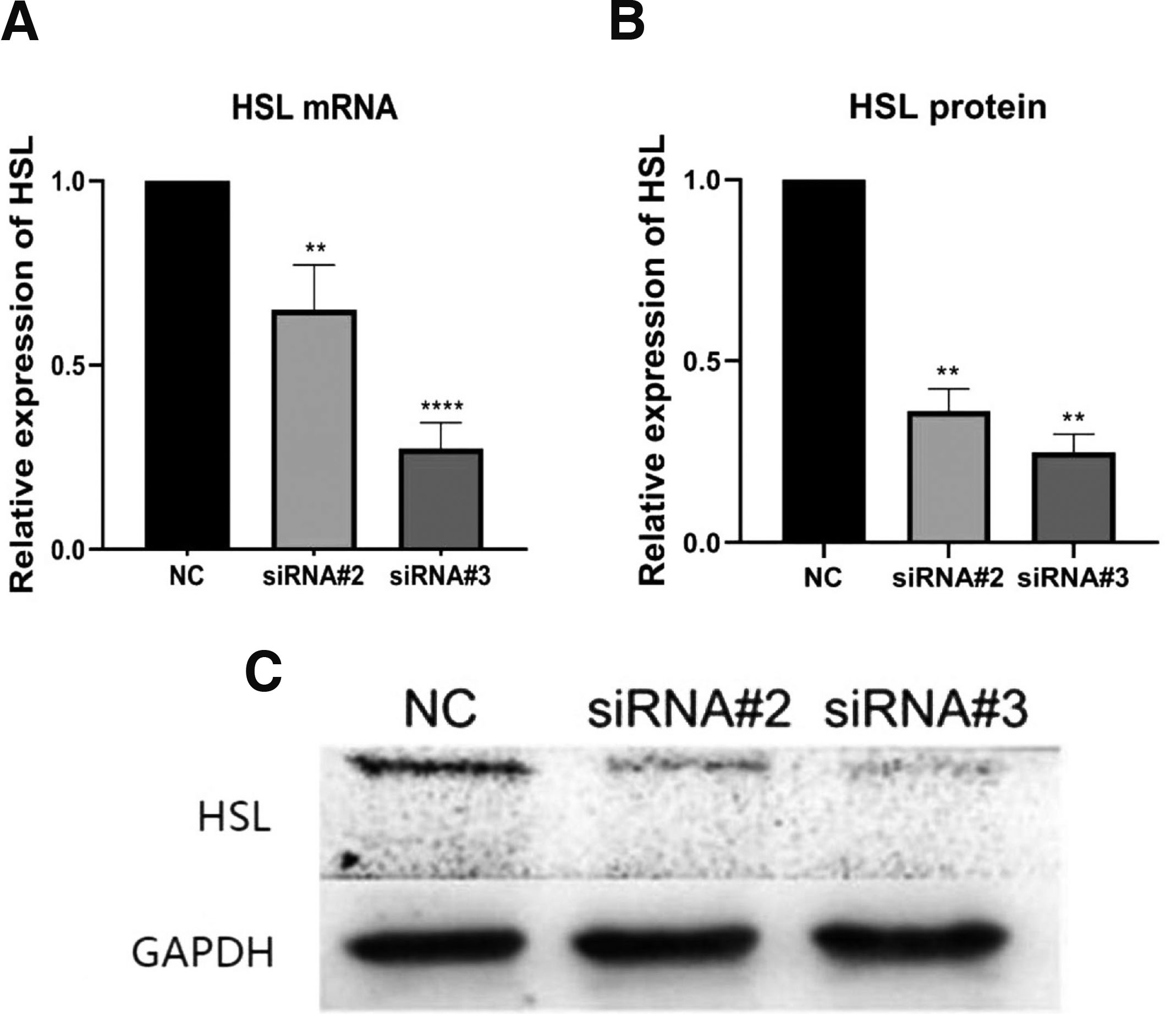
To verify whether the designed siRNA silenced the HSL gene in the atRA group, we determined the effects of the two chemosynthetic siRNAs (#2 and #3) in MEPM cells after transient transfection. The two siRNAs against HSL reduced mRNA and protein expression, and cells transfected with siRNA#3 showed the most significantly reduced mRNA and protein levels of HSL (Fig. 4). Therefore, siRNAs #2 and #3 were selected to study the function of HSL.
Fig. 4. RNAi knockdown of HSL in MEPM cells.
(A) qRT-PCR was used to detect the effect of HSL RNAi on the mRNA expression of HSL. (B) The protein level of HSL in MEPM cells was detected by western blot. (C) Quantitative densitometry evaluation of HSL was shown. **P<0.01, ****P<0.0001 versus the negative control groups. HSL, hormone-sensitive lipase; MEPM, mouse embryonic palatal mesenchyme; atRA, all-trans retinoic acid; NC, negative control.
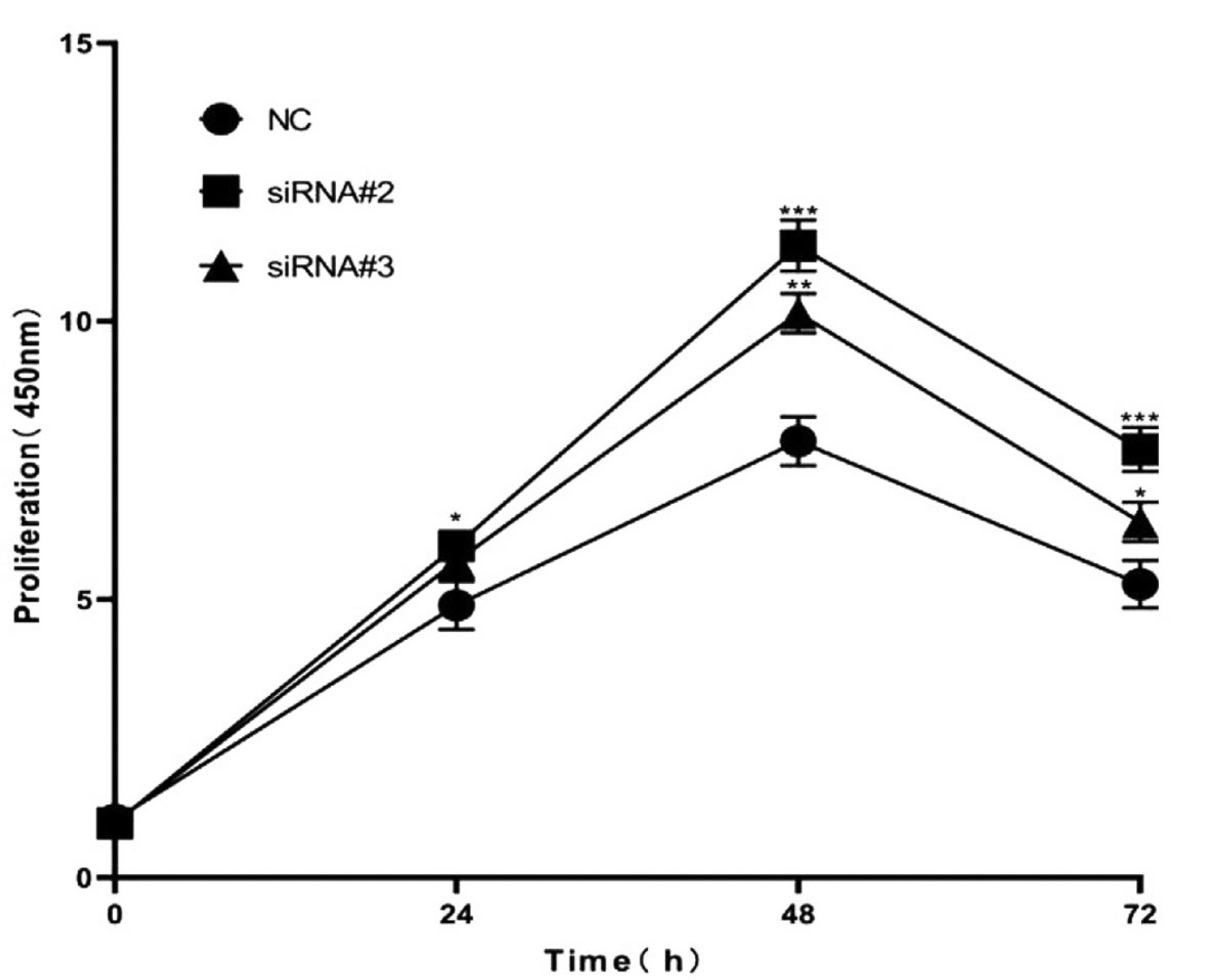
Promotion of cell proliferation by HSL siRNA
In the atRA group, the CCK-8 assay was used to assess the effect of HSL siRNA on the proliferation of MEPM cells cultured in 96-well plates. After 48 h of transfection, the HSL siRNA-transfected cells showed an increase in proliferation compared to the negative siRNA-transfected cells (Fig. 5).
Fig. 5. The downregulation of HSL expression promoted the proliferation ability of MEPM cells.
MEPM cells transfected with negative control and HSL siRNAs were used for the CCK8 assay. Cell viability in HSL siRNA-transfected cells was significantly increased compared with corresponding controls. *P<0.05, **P<0.01, ***P<0.001 versus the control groups. HSL, hormone-sensitive lipase; MEPM, mouse embryonic palatal mesenchyme; atRA, all-trans retinoic acid; NC, negative control.
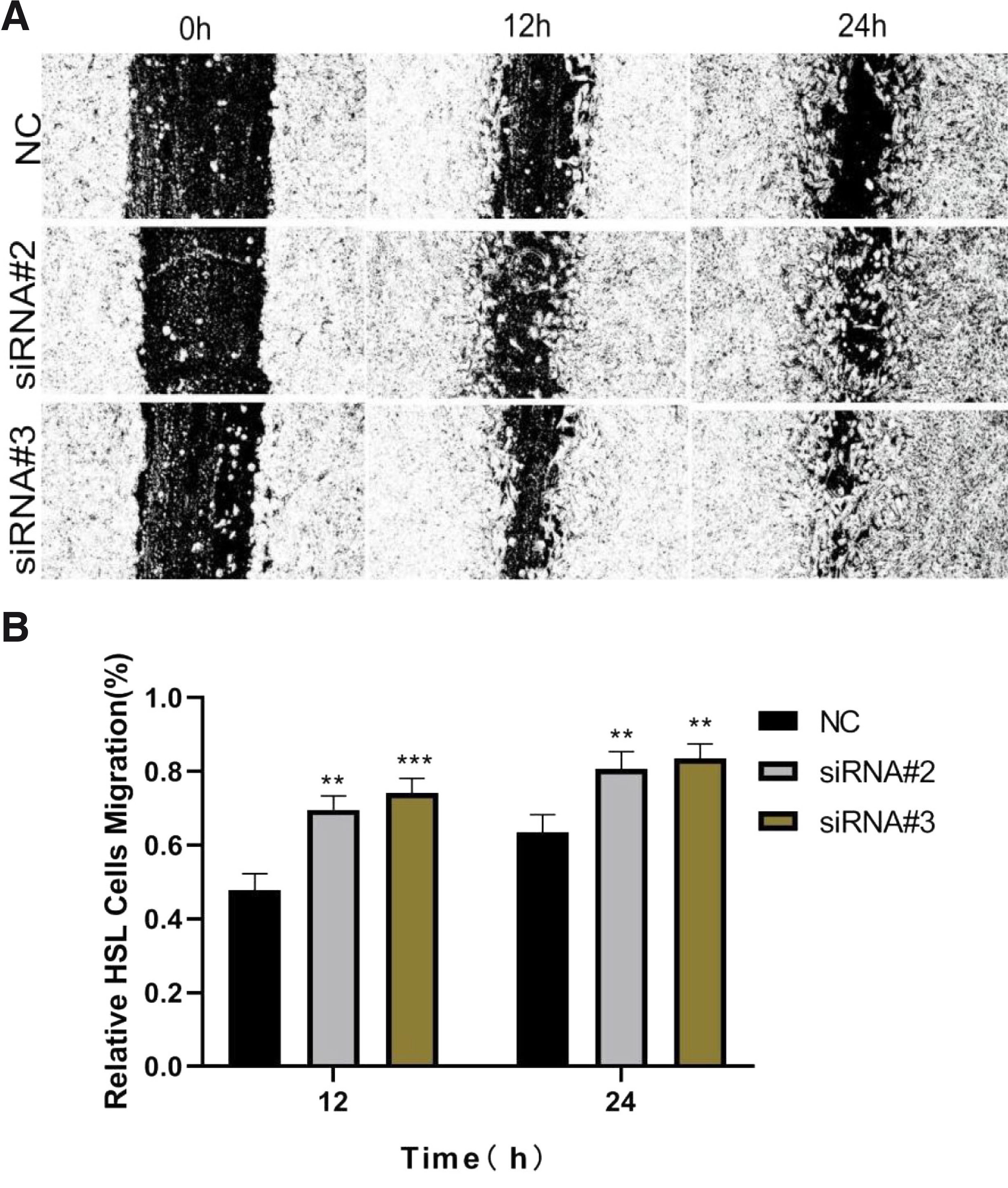
Promotion of cell migration by HSL siRNA
In the atRA group, we studied the influence of HSL knockdown on MEPM cell migration capacity by the wound scratch assay. After 48 h of transfection, the migration of cells transfected with HSL siRNA was significantly increased compared with that of the negative siRNA-transfected cells (Fig. 6).
Fig. 6. Knockdown of HSL promoted MEPM cells migration as determined by the scratch wound assay.
Representative photomicrographs of MEPM cell migration were taken, and an inverted fluorescence microscope measured the relative distances of migration. (A) The typical photomicrographs of MEPM cells’ migration. (B) The results showed that the relative mobility of cells in the siRNA group was significantly higher than that in the negative control group. Data were expressed as mean±SD from three separate experiments. **P<0.01, ***P<0.001 versus the negative control. Original magnification 100×. HSL, hormone-sensitive lipase; MEPM, mouse embryonic palatal mesenchyme; atRA, all-trans retinoic acid; NC, negative control.
Discussion
Cleft lip and palate is the most prevalent congenital orofacial malformation, occurring in approximately 1:700 live human births worldwide each year (Salari et al., 2022). Therefore, it is of significant clinical importance to explore the pathogenesis of cleft lip and palate. Since morphological events in mice closely resemble those seen in humans, mouse genetic models with palatal defects have been valuable for analyzing the genes that regulate palatogenesis and determining the genetic causes of human cleft palate (Oliver et al., 2021).
atRA plays an essential role in regulating morphogenesis, cell differentiation, and proliferation during embryonic development (Clagett-Dame et al., 2002). Many animal models and cytotoxicity tests have found that atRA is widely used as a teratogen to determine the pathogenesis of cleft palate (Han et al., 2006; Zhang et al., 2017a). Cleft palate occurred in mice and humans when exposed to both deficiency and overdoses of atRA (Ackermans et al., 2011; Wilcox et al., 2007). Previous studies have confirmed that embryos exposed to atRA (70 mg/kg) at E10.0 show a higher incidence of cleft palate, the palatal shelves of which become growth retarded and fail to contact one another (Abbott et al., 1990; Shu et al., 2019). In this study, we observed that the palatal shelves were abnormally small and dysplastic. The results indicated that palatal shelf growth was permanently impeded in atRA-exposed embryos on gestation Day 15.5.
HSL is a key enzyme in regulating lipid metabolism and is highly expressed in adipose tissue (Pajed et al., 2021). Previous studies confirmed that a defect in lipolysis was detected in the cleft palate of rats (Stefanovich et al., 1971), and modulation of lipid metabolic defects rescued the cleft palate in gene mutant mice (Iwata et al., 2014). These results indicated that lipid metabolism plays a significant role in normal physiological function and development, particularly in the maxillofacial region and palate. During embryonic development, mesenchymal cell proliferation is the main process in shelf outgrowth, and cleft palate occurs when this process is disrupted (Zhang et al., 2017b). However, whether HSL is present in MEPM cells and the mechanism of HSL in cleft palates remain unclear. The present study showed that HSL was strongly expressed at E15.5 in cleft palate MEPM cells induced by atRA, while it was downregulated in the typical embryonic palate. These observations indicated that HSL is involved in the development of MEPM cells. Increased HSL may result in alterations in lipolysis activities and other lipid metabolic pathways, causing reduced mesenchymal cell proliferation.
MEPM cell migration and proliferation play significant roles in forming facial structures (He et al., 2008). Many studies have investigated whether atRA inhibits MEPM cell proliferation (Liu et al., 2016; Liu et al., 2021; Yoshioka et al., 2021; Zhang et al., 2017b). However, the molecular mechanism by which HSL affects cell migration and proliferation needs to be fully clarified. In the present study, the CCK-8 assay results showed that the knockdown of HSL by siRNA promoted MEPM cell proliferation in the atRA group. Furthermore, HSL siRNA significantly promoted the migration of MEPM cells in the atRA group. We provide evidence for the first time that HSL contributes to altering MEPM cell migration and proliferation. Our data suggest that the promotion of cell proliferation and migration induced by HSL siRNA may be related to the development of the palatal shelf in mice.
In summary, this study confirmed that HSL is involved in cleft palate induced by atRA, and the expression of HSL was upregulated in the atRA group. The downregulation of HSL using RNAi promotes the proliferation and migration of MEPM cells in the atRA group. Therefore, key lipolytic enzymes may affect the development of the palatal shelves, as atRA enhanced HSL levels to inhibit embryonic palate growth. Our study had several limitations. First, we investigated the function of HSL in the cleft palate at E15.5; thus, it is yet to be known whether our results apply to other stages of palatogenesis.
Furthermore, we examined a simple gene in a very complex signaling pathway. Further studies should be applied to verify the relationship between HSL and lipid metabolism-related pathways in MEPM cell migration and proliferation and to determine whether the increase in cellular HSL levels may contribute to alterations of lipid metabolic activity in the palatal mesenchyme. Our findings provide new insights into the underlying mechanism of cleft palate formation. They may help identify new candidate markers for prenatal screening for cleft palate and new targets for its diagnosis and treatment.
Material and Methods
Animals
Laboratory Animal Ethical Committee of the Medical College of Shantou University (Shantou, China) approved the study protocol involving animals. Sixty Kunming mice (20 males and 40 females, aged 8–10 weeks) were purchased from the Shantou University Laboratory Animal Technology Co., Ltd. (Shantou, China). Females were mated with fertile males with similar weights and ages overnight. The presence of a vaginal plug the following morning was considered indicative of embryonic gestation Day 0.5 (E0.5). We obtained a total of 22 pregnant females, and they were randomly divided into two oral gavage groups at E10.5: atRA-treated groups received 70 mg/kg atRA (Sigma; USA) dissolved in corn oil, and an equivalent volume of corn oil alone was given to the control group. At E15.5, pregnant mice were sacrificed by cervical dislocation. The fetuses were removed from the uterus, and 108 embryos were examined.
Tissue fixation and section preparation
Seventy embryonic heads were dehydrated and greased using an ascending ethanol series and xylene after spending the night in 4% paraformaldehyde (PFA). Then, the embryonic heads were embedded in paraffin and processed into 5 μm-thick serial coronal sections of the fetal heads.
Histological observation
Hematoxylin and eosin (H&E) staining was performed on coronal sections of the embryonic heads to evaluate changes in palatal shelves during palatogenesis according to standard procedures. The results were compared between the control and atRA-treated groups.
Immunohistochemistry (IHC)
The coronal sections were exposed to microwave irradiation for 30 min by applying citric acid buffer (pH 6.0) at 95 °C for antigen activation. Normal donkey serum (10% v/v in PBS, Vector) was used to block nonspecific staining for one hour. Following optimization, the HSL primary antibody (1:50; 17333-1-AP; Proteintech; China) was used at 4 °C overnight. The sections were washed using phosphate-buffered saline (PBS; Gibco; USA) and incubated with secondary antibodies for two hours at room temperature. Staining was visualized following incubation with 3,3′-diaminobenzidine (DAB; OriGene Technologies, Inc.; USA). The emergence of brown granules was regarded to indicate positive staining. Sections were counterstained with hematoxylin for two seconds. Images were obtained with a Zeiss Axioskop 40 (Leica Microsystems; Germany).
Cell culture and treatment
Pregnant mice were euthanized at E15.5, and the fetuses were collected. Only the palate mucosa of the fetal mice was selectively isolated under a microscope and digested with dispase II (Godo Shusei Co.; Japan) and trypsin EDTA (Biological Industries, Israel) at 37 °C for 30 min. After centrifugation, mouse embryonic palatal mesenchymal (MEPM) cells were isolated using a cell strainer with a pore size of 70 µm (BD Falcon; USA). The cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; USA) containing 10% fetal bovine serum (FBS; Gibco; USA) and 1% penicillin/streptomycin (Sigma; Germany) in an atmosphere with 5% CO2 at 37 °C. The medium was replaced every 2-3 days and then subcultured at the exponential growth stage (80% confluence).
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated with TRIzol from MEPM cells or transfected cells (Sigma; Germany). cDNA was synthesized using the PrimeScript RT reagent kit (TaKaRa; Japan), and the qRT-PCR mix was prepared with the SYBR Premix Ex Taq kit (Takara; Japan). Then, qRT-PCR was performed using the SLAN-96S qPCR system (Hongshi; China).
Table 1
List of primers used in this study
| Primer Name | Primer Sequence (5´-3´) F=Forward, R=Reverse |
|---|---|
| HSL gene | F=5´-TGAGATGGTAACTGTGAGCC-3´ |
| R=5´-ACTGAGATTGAGGTGCTGTC-3´ | |
| GAPDH | F=5´-AGAACATCATCCCTGCCTCTACTG-3´ |
| R=5´-AAATGAGCTTGACAAAGTGGTCGT-3´ |
Western blot (WB) analysis
All lysates were prepared from MEPM cells or transfected cells using 2 × SDS lysis buffer supplemented with protease inhibitors (M250, Amresco, Ohio, United States) and phosphate inhibitors (WB0117, Weiao Biotech, China). Protein content was determined using a standard BCA protein assay kit (Dingguo, Beijing, China); 50 μg of protein was fractionated by 12% SDS-PAGE and electroblotted to nitrocellulose membranes. After blocking with 5% nonfat milk, the membranes were incubated with anti-HSL (1:1000; 17333-1-AP; Proteintech; USA) and anti-GAPDH (1:5000; 10494-1-AP; Proteintech; USA) primary antibodies at 4 °C and secondary antibodies (1:10000; SA00001-2; Proteintech; USA) for one hour at room temperature. Blots were created using the enhanced chemiluminescence reagent (Beyotime; China), and ImageJ software was used to assess band intensities (NIH, Bethesda; USA).
Transfection with small interfering RNAs (siRNA)
atRA-treated group cells were cultured in 6-well plates (1×104 cells/well) in DMEM containing 10% FBS. According to the manufacturer's instructions, the MEPM cells were transfected with a final concentration of 70 pmol HSL siRNA (Huzhou HippoBio Co., Ltd.; China) using Lipofectamine RNAiMAX Reagent (Invitrogen; USA) for 48 h. A negative siRNA (Huzhou HippoBio Co., Ltd.; China) was used as the control group. These cells were collected to confirm whether the designed siRNA silenced the genes. The effectiveness of the target gene knockdown was determined using qRT-PCR and WB.
Cell counting kit-8 (CCK8) assay
atRA-treated group cells (5×103 cells/well) were cultured in a 96-well plate (Corning; USA) in DMEM containing 10% FBS at 37 °C. Following 24, 48, and 72 h, incubation with 10 µl CCK8 solution (Beyotime Biotechnology; China) at 37 °C for an additional 2 hours. The absorbance at 450 nm was measured using a microplate reader.
Wound healing assays
atRA-treated group cells (5×105cells/well) were collected and seeded into 12-well plates. After transfection for 48 h, cells were starved for 12 h. The DMEM was replaced with a medium containing 2.5% FBS. A line-shaped scratch was made by passing a pipette tip after the cells merged into the monolayer cells. The width between the borders of the scratches was photographed in each group at 0, 12, and 24 h using an inverted microscope connected to a digital camera (Olympus Corporation; Japan).
Statistical analysis
Data for each parameter were obtained in triplicate. All data were evaluated and analyzed for significance by SPSS version 22.0. Comparisons between groups were performed using one-way analysis of variance (ANOVA) or Student’s t-test. P < 0.05 was deemed a statistically significant difference.
Declarations
Competing interests
The authors declare that no competing interests exist.
References
Abbott B. D., Birnbaum L. S. (1990). Retinoic acid-induced alterations in the expression of growth factors in embryonic mouse palatal shelves. Teratology 42: 597-610.
Abramyan J., Richman J. M. (2015). Recent insights into the morphological diversity in the amniote primary and secondary palates. Developmental Dynamics 244: 1457-1468.
Ackermans M. M.G., Zhou H., Carels C. E.L., Wagener F. A.D.T.G., Von den Hoff J. W. (2011). Vitamin A and clefting: putative biological mechanisms. Nutrition Reviews 69: 613-624.
Al Tanoury Z., Piskunov A., Rochette-Egly C. (2013). Vitamin A and retinoid signaling: genomic and nongenomic effects. Journal of Lipid Research 54: 1761-1775.
Alois C. I., Ruotolo R. A. (2020). An overview of cleft lip and palate. Journal of the American Academy of Physician Assistants 33: 17-20.
Bush J. O., Jiang R. (2012). Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139: 231-243.
Clagett-Dame M., DeLuca H. F. (2002). T HE R OLE OF V ITAMIN A IN M AMMALIAN R EPRODUCTION AND E MBRYONIC D EVELOPMENT . Annual Review of Nutrition 22: 347-381.
Gao L., Wang Y., Yao Y., Zhang G., Han G., Wu W. (2016). 2,3,7,8-Tetrachlorodibenzo- p -dioxin Mediated Cleft palate by Mouse Embryonic Palate Mesenchymal Cells. Archives of Oral Biology 71: 150-154.
Han J., Xiao Y., Lin J. X., Zhang Z. F., Li Y. (2006). Establishment and application of fetal rolling palate organ culture model of mouse. Wei Sheng Yan Jiu 35: 33-35.
He F., Xiong W., Yu X., Espinoza-Lewis R., Liu C., Gu S., Nishita M., Suzuki K., Yamada G., Minami Y., Chen Y.P. (2008). Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development 135: 3871-3879.
Iwata J., Suzuki A., Pelikan R. C., Ho T.V., Sanchez-Lara P. A., Chai Y. (2014). Modulation of lipid metabolic defects rescues cleft palate in Tgfbr2 mutant mice. Human Molecular Genetics 23: 182-193.
Knittel C. H., Devaraj N. K. (2022). Bioconjugation Strategies for Revealing the Roles of Lipids in Living Cells. Accounts of Chemical Research 55: 3099-3109.
Liu X., Qi J., Tao Y., Zhang H., Yin J., Ji M., Gao Z., Li Z., Li N., Yu Z. (2016). Correlation of proliferation, TGF-β3 promoter methylation, and Smad signaling in MEPM cells during the development of ATRA-induced cleft palate. Reproductive Toxicology 61: 1-9.
Liu X., Zhang Y., Shen L., He Z., Chen Y., Li N., Zhang X., Zhang T., Gao S., Yue H., Li Z., Yu Z. (2021). LncRNA Meg3-mediated regulation of the Smad pathway in atRA-induced cleft palate. Toxicology Letters 341: 51-58.
Mossey P. A., Little J., Munger R. G., Dixon M. J., Shaw W. C. (2009). Cleft lip and palate. The Lancet 374: 1773-1785.
Nohara A., Owaki N., Matsubayashi J., Katsube M., Imai H., Yoneyama A., Yamada S., Kanahashi T., Takakuwa T. (2022). Morphometric analysis of secondary palate development in human embryos. Journal of Anatomy 241: 1287-1302.
Oliver J. D., Jia S., Halpern L. R., Graham E. M., Turner E. C., Colombo J. S., Grainger D. W., D'Souza R. N. (2021). Innovative Molecular and Cellular Therapeutics in Cleft Palate Tissue Engineering. Tissue Engineering Part B: Reviews 27: 215-237.
Pajed L., Taschler U., Tilp A., Hofer P., Kotzbeck P., Kolleritsch S., Radner F. P. W., Pototschnig I., Wagner C., Schratter M., Eder S., Huetter S., Schreiber R., Haemmerle G., Eichmann T. O., Schweiger M., Hoefler G., Kershaw E. E., Lass A., Schoiswohl G. (2021). Advanced lipodystrophy reverses fatty liver in mice lacking adipocyte hormone-sensitive lipase. Communications Biology 4: 323.
Salari N., Darvishi N., Heydari M., Bokaee S., Darvishi F., Mohammadi M. (2022). Global prevalence of cleft palate, cleft lip and cleft palate and lip: A comprehensive systematic review and meta-analysis. Journal of Stomatology, Oral and Maxillofacial Surgery 123: 110-120.
Shu X., Dong Z., Cheng L., Shu S. (2019). DNA hypermethylation of Fgf16 and Tbx22 associated with cleft palate during palatal fusion. Journal of Applied Oral Science 27: e20180649.
Stefanovich V., Gianelly A. (1971). Preliminary Studies of the Lipids of Normal and Cleft Palates of the Rat. Journal of Dental Research 50: 1360-1360.
Suzuki A., Abdallah N., Gajera M., Jun G., Jia P., Zhao Z., Iwata J. (2018). Genes and microRNAs associated with mouse cleft palate: A systematic review and bioinformatics analysis. Mechanisms of Development 150: 21-27.
Sztrolovics R., Wang S. P., Lapierre P., Chen H. S., Robert M. F., Mitchell G. A. (1997). Hormone-sensitive lipase (Lipe): Sequence analysis of the 129Sv mouse Lipe gene. Mammalian Genome 8: 86-89.
Wang S. P., Wu J. W., Bourdages H., Lefebvre J. F., Casavant S., Leavitt B. R., Labuda D., Trasler J., Smith C. E., Hermo L., Mitchell G. A. (2014). The Catalytic Function of Hormone-Sensitive Lipase is Essential for Fertility in Male Mice. Endocrinology 155: 3047-3053.
Wang W., Jian Y., Cai B., Wang M., Chen M., Huang H. (2017). All-Trans Retinoic Acid-Induced Craniofacial Malformation Model: A Prenatal and Postnatal Morphological Analysis. The Cleft Palate-Craniofacial Journal 54: 391-399.
Wilcox A. J., Lie R. T., Solvoll K., Taylor J., McConnaughey D. R., Åbyholm F., Vindenes H., Vollset S. E., Drevon C. A. (2007). Folic acid supplements and risk of facial clefts: national population based case-control study. BMJ 334: 464.
Worley M. L., Patel K. G., Kilpatrick L. A. (2018). Cleft Lip and Palate. Clinics in Perinatology 45: 661-678.
Yoshioka H., Mikami Y., Ramakrishnan S. S., Suzuki A., Iwata J. (2021). MicroRNA-124-3p Plays a Crucial Role in Cleft Palate Induced by Retinoic Acid. Frontiers in Cell and Developmental Biology 9: 621045.
Zhang H., Liu X., Gao Z., Li Z., Yu Z., Yin J., Tao Y., Cui L. (2017a). Excessive retinoic acid inhibit mouse embryonic palate mesenchymal cell growth through involvement of Smad signaling. Animal Cells and Systems 21: 31-36.
Zhang Y., Dong S., Wang J., Wang M., Chen M., Huang H. (2017b). Involvement of Notch2 in all-trans retinoic acid-induced inhibition of mouse embryonic palate mesenchymal cell proliferation. Molecular Medicine Reports 16: 2538-2546.
Zhou H., zhang J., Yan Z.K., Qu M., Zhang G., Han J., Wang F., Sun K., Wang L., Yang X. (2022). DECR1 directly activates HSL to promote lipolysis in cervical cancer cells. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1867: 159090.
Zhou J., Xu B., Shi B., Huang J., He W., Lu S., Lu J., Xiao L., Li W. (2011). A Metabonomic Approach to Analyze the Dexamethasone-Induced Cleft Palate in Mice. Journal of Biomedicine and Biotechnology 2011: 1-8.