Int. J. Dev. Biol. 66: 235 - 241 (2022)
Special Issue: Developmental Biology in Greece
Evidence of Swim secretion and association with extracellular matrix in the Drosophila embryo
Open Access | Original Article | Published: 22 November 2021
Abstract
Secreted wingless-interacting protein (Swim) is the Drosophila ortholog gene of the mammalian Tubulointerstitial Nephritis Antigen like 1 (TINAGL1), also known as lipocalin-7 (LCN7), or adrenocortical zonation factor 1 (AZ-1). Swim and TINAGL1 proteins share a significant homology, including the somatomedin B and the predictive inactive C1 cysteine peptidase domains. In mammals, both TINAGL1 and its closely related homolog TINAG have been identified in basement membranes, where they may function as modulators of integrin-mediated adhesion. In Drosophila, Swim was initially identified in the eggshell matrix and was subsequently detected in the culture medium of S2 cells. Further biochemical analysis indicated that Swim binds to wingless (wg) in a lipid-dependent manner. This observation, together with RNAi-knockdown studies, suggested that Swim is an essential cofactor of wg-signalling. However, recent elegant genetic studies ruled out the possibility that Swim is required alone to facilitate wg-signalling in Drosophila, because flies without Swim are viable and fertile. Here, we use the UAS/Gal4 expression system together with confocal imaging to analyze the in vivo localization of a chimeric Swim-GFP in the developing Drosophila embryo. Our data fully support the notion that Swim is an extracellular matrix component that is secreted upon ectopic expression and preferentially associates with the basement membranes of various organs and with the specialized tendon matrix at the muscle attachment sites (MAS). Interestingly, the accumulation of Swim at the MAS does not require integrins. In conclusion, Swim is an extracellular matrix component, and Swim may exhibit overlapping functions in concert with other undefined components.
Keywords
basement membrane, muscle attachment sites, tubulointerstitial nephritis antigen like 1 (TINAGL1), lipocalin-7 (LCN7), integrins
Introduction
Extracellular matrix (ECM) is a meshwork of interacting secreted proteins that fills the space between cells and provides the structural framework to hold tissues together. ECM not only functions as cellular glue in tissue formation but also facilitates the interaction and presentation of various extracellular growth factors to their plasma membrane receptors (Hynes, 2009). ECM molecular composition determines the physical and chemical properties of the various matrices within the body, which significantly differ among organs. For instance, basement membranes encircle internal organs and provide support for tissue growth and differentiation, or serve as a track for migrating cells (Cruz Walma and Yamada, 2020). In addition, the interstitial matrix provides more specialized functions, including the strong fibers of tendons or the fibrous capsules around organs. The diverse array of ECM functions is largely influenced by the organization of the structural components, which are often modified by specialized proteins with or without enzymatic properties. Thus, increased insight into the molecular mechanisms of ECM functions requires the characterization of all matricellular proteins, which is currently an emerging research theme in matrix biology (Hynes and Naba, 2012; Bonnans et. al., 2014).
The vertebrate tubulointerstitial nephritis antigen (TINAG) and the tubulointerstitial nephritis antigen like 1(TINAGL1), or adrenocortical zonation factor 1 (AZ-1), or lipocalin 7, compose a family of closely related proteins, containing a somatomedin-B like motif followed by a peptidase C1A-C domain (Kalfa et al., 1994; Brömme et al., 2000). Detailed analysis of the TINAGL1 protein sequence showed that the peptidase domain contains key amino acid substitutions within the catalytic site, which likely renders it inactive (Wex et al., 2001). TINAGL1 has been classified as a matricellular protein because several lines of evidence support its ability to associate with the basement membrane and to interact with fibronectin and laminin, both of which are structural components of the ECM (Igarashi et al., 2009; Hynes and Naba, 2012).
Human TINAGL1 is expressed in vascular smooth cells, aorta, heart, placenta, skeletal muscle and kidney (Wex et al., 2001). Additional studies have shown that mouse TINAGL1 is expressed in the blastomere and morulae embryos (Sakurai et al., 2014), while before implantation TINAGL1 is expressed in the blastocyst trophectoderm. Post-implantation, TINAGL1 expression is restricted to the extraembryonic regions and the Reichert membrane (Igarashi et al., 2009). Interestingly, a recent study in LM2 cell line demonstrated that TINAGL1 modulates FAK and EGFR mediated signalling pathways, by antagonizing fibronectin binding to integrins α5β1 and αvβ1 and additionally inhibiting epidermal growth factor receptor dimerization. Consequently, TINAGL1 suppresses tumour growth in the triple-negative breast cancer (TNBC) model (Shen et al., 2019). These data were consistent with earlier studies that demonstrated TINAGL1 secretion is regulated by Sec23a and, interestingly, had a metastatic suppressor effect (Korpal et al., 2011). The therapeutic potential of TINAGL1 is currently explored by directly targeting its expression in the tumour microenvironment (Musetti and Huang, 2021). Interestingly, mice without TINAGL1 are viable, but exhibit impaired fertility (Takahashi et al., 2016).
In Drosophila, the homolog gene CG3074 is closely related to TINAGL1, rather than to the TINAG. The fly homolog displays structural similarities to its vertebrate counterpart, while initial studies identified its deposition in the anterior part of the eggshell matrix (Fakhouri et al., 2006). The CG3074 was renamed as secreted wg-interacting molecule (swim) because it was identified in the S2 cell culture medium and could bind to wg in a lipid-dependent manner (Mulligan et al., 2012). Additional knock-down studies of swim suggested its role as a cofactor that facilitates long-range wg signalling (Mulligan et al., 2012). However, recent evidence shows that null mutants for Swim are viable and fertile and therefore Swim function is either masked by genetic redundancy with other wg co-factors or does not participate in wg-signalling at all (McGough et al., 2020).
Our present work showed that Swim is a secreted protein, and identified its extracellular distribution properties in the developing Drosophila embryo. We used the UAS/Gal4 inducible expression system and found that a chimeric Swim-GFP expressed ectopically in muscle or gut epithelial cells is secreted in the hemocoel and deposited in the basement membranes surrounding the gut, the nervous system and the specialized tendon matrix at the muscle attachment sites (MAS). Our data provide an alternative hypothesis for a putative redundant function for Swim in the ECM remodelling in tissue growth and maintenance in Drosophila.
Results and Discussion
Generation of UAS: Swim-GFP
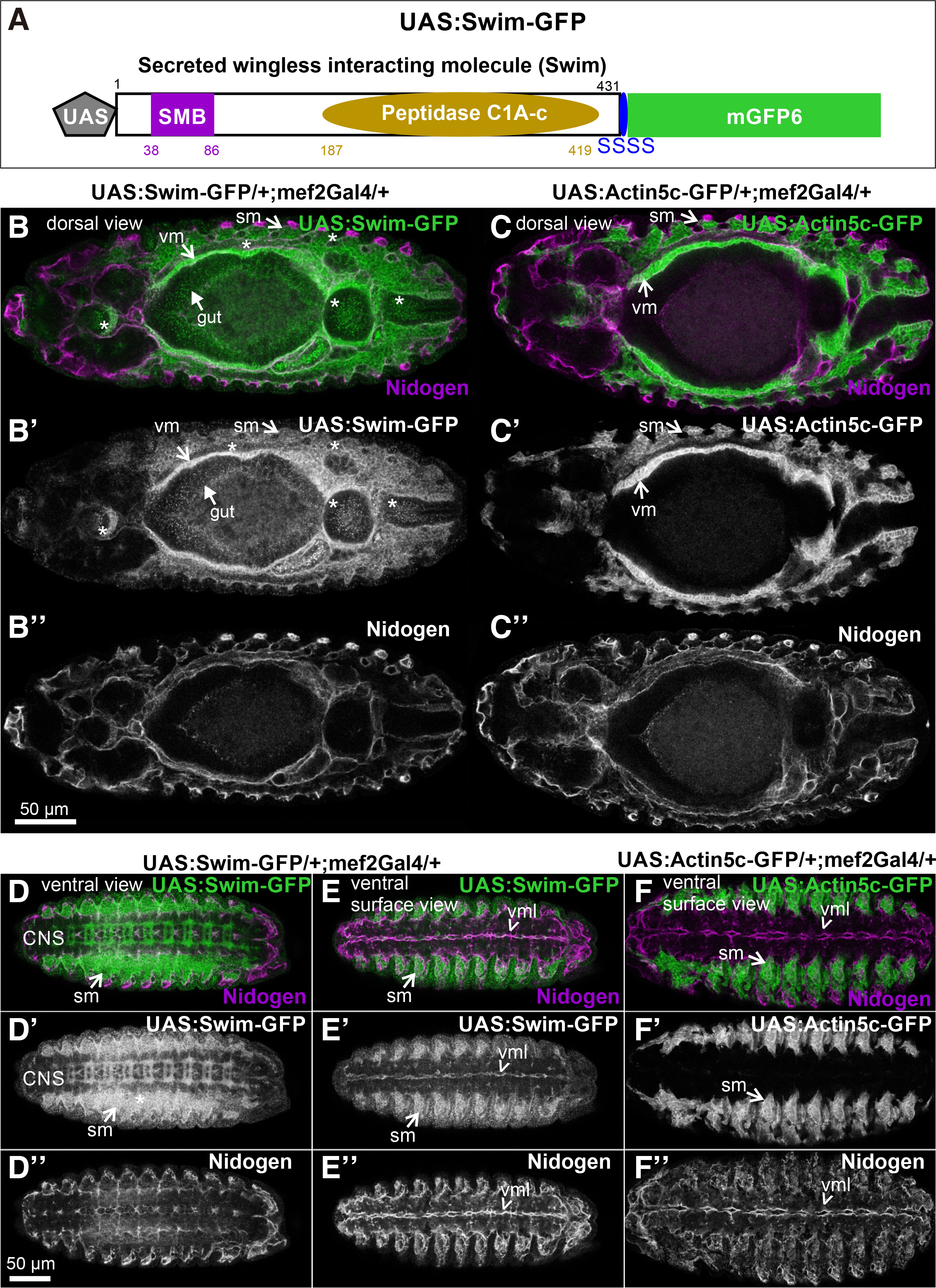
Swim shares 41% identity and 55% similarity with the human TINAGL1 across the entire protein sequence. InterPro scan analysis (www.ebi.ac.uk/interpro) predicted two conserved domains: the Somatomedin-B-domain (aa: 38-86) and the peptidase C1A-C domain (aa:187-419). To investigate the localization properties of secreted wingless-interacting protein (swim) in vivo, we first engineered a Swim-GFP fusion gene under the control of a UAS promoter, allowing expression of the fluorescently-tagged protein in different tissues using the GAL4 system (Brand and Perrimon, 1993). Specifically, we fused at the C-terminus of swim (its open reading frame) a linker of four serines, followed by the coding sequence of mGFP6 (Fig. 1A). This approach allowed us to follow the expression and subcellular localization of the Swim-GFP in both living and fixed embryos.
Fig. 1. Generation and inducible expression of UAS: Swim-GFP in Drosophila embryos.
(A) Schematic representation of the engineered Swim-GFP fusion gene under the control of UAS promoter. (B-F) Stage 13 embryos probed with an antibody against Nidogen (magenta), expressing (B-B”, D-D”, E-E”) Swim-GFP (green in B, D, E and white in B’, D’, E’), or (C-C”, F-F”) Actin5c-GFP (green in C, F and white in C’, F’) both driven by mef2Gal4. Dorsal view of (B, B’) Swim-GFP distribution reveals heavy accumulation within both the developing visceral and somatic musculature and their surrounding extracellular space. Asterisks (*) indicate the diffused pattern of Swim-GFP dispersion in the developing head, gut and posterior embryonic areas, which is strictly unique to Swim-GFP and absent when compared to (C, C’) Actin5c-GFP distribution in the relevant area. (B, B”) Nidogen localization partially coincides with the extracellular distribution of Swim-GFP distribution, however appears distinct to (C, C”) Actin5c-GFP detected pattern. Ventral view of (D, D’ and E, E’) Swim-GFP compared to (F-F’) Actin5c-GFP distribution demonstrates Swim-GFP secretion and deposition in the developing neuropile and its colocalization with (D, D”, E, E” and F, F”) Nidogen merely in the midline of the embryonic ventral surface area. Each image is representative of at least five different imaged embryos of the same genotype and markers used. CNS, central nervous system; sm, somatic muscles; vm, visceral muscles; vml, ventral midline Scale bars: 50 μm in all panels.
Swim-GFP expression and distribution in Drosophila embryos
To identify whether Swim-GFP was secreted, we selected three Gal4 drivers that are expressed in a distinct pattern and tissues: a) mef2Gal4 is expressed in the somatic and visceral muscles and also in the dorsal vessel at the late stage of embryogenesis; b) 48YGal4 is expressed in the entire intestine; and c) drmGal4 is expressed in the hindgut and malpighian tubules but also at lower levels in the midgut, proventriculus and in tendon cells.
First, we analyzed the extracellular localization properties of Swim-GFP by examining embryos at stage 13. Expression of Swim-GFP in the developing muscles using the mef2Gal4 revealed that Swim-GFP was not only distributed within the somatic and visceral muscles but also was heavily accumulated in a diffused pattern in the extracellular space around the muscle tissues (Fig. 1 B-B’). By contrast, actin5c-GFP was strictly accumulated within the muscle tissue (Fig. 1 C-C’). To verify whether the extracellular distribution of Swim-GFP is associated with embryonic basement membranes, we stained embryos with antibodies against nidogen (Dai et al., 2018; Wolfstetter et al., 2019). The extracellular distribution of Swim-GFP partially coincided with nidogen in certain areas surrounding the somatic muscles and the visceral muscles (Fig. 1 B,B’’). However, we observed Swim-GFP in a punctuated pattern in the head region, along the gut region, and in the posterior part of the embryo (Fig. 1B’). Because the UAS/Gal4 system achieves relatively high levels of expression, it is possible that the high amount of secreted Swim-GFP is gradually incorporated during the basement membrane assembly.
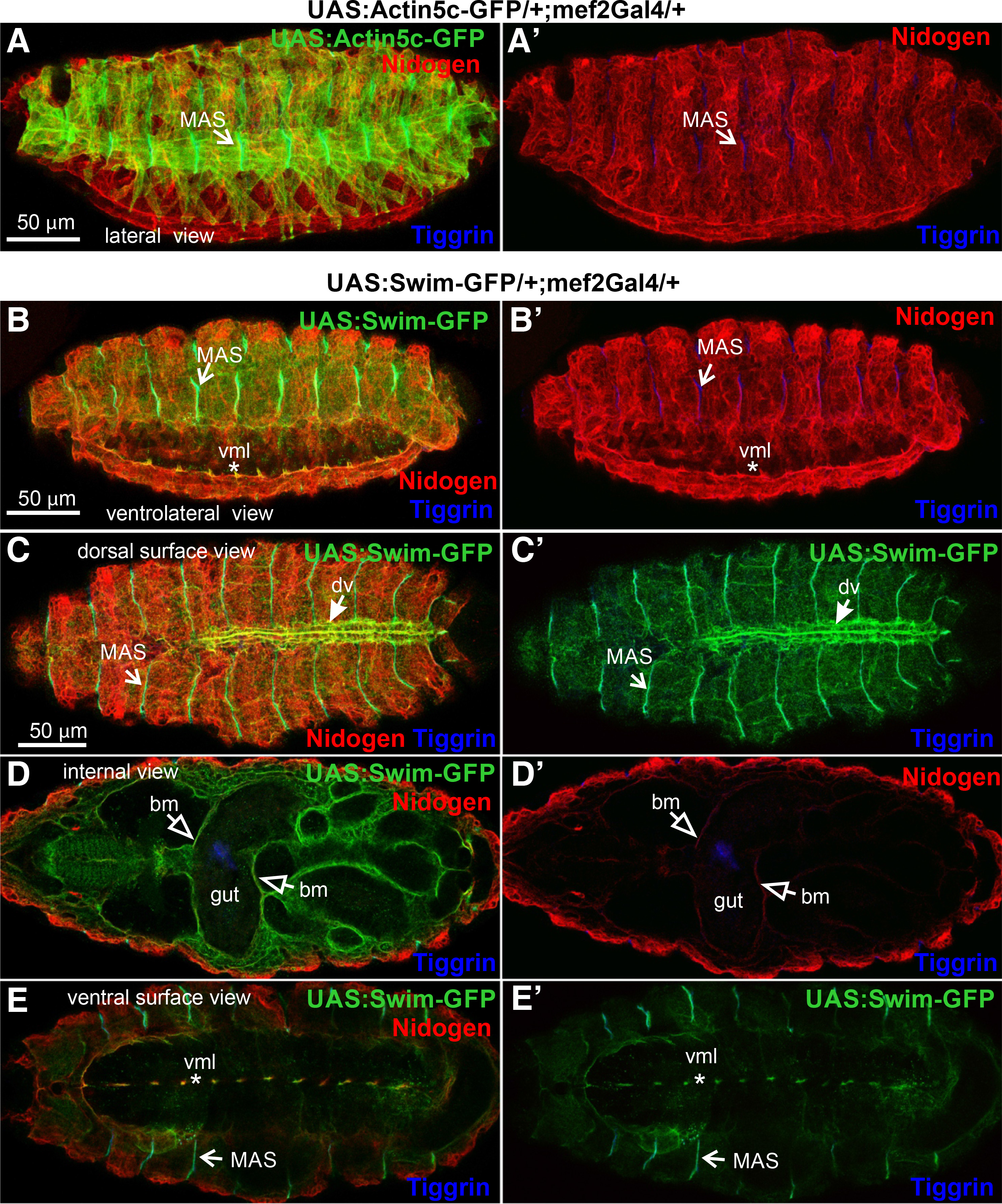
We further examined the distribution of Swim-GFP in the ventral side of the embryo during ventral nerve cord formation, at which stage the basement membrane has not been fully assembled (Broadie et al., 2011). We found an intense deposition of Swim-GFP in the developing neuropile (Fig. 1 D-D’’, E-E’’). However, Swim-GFP was colocalized with nidogen only in the midline of the ventral surface area of the embryo (Fig. 1. D’’-E’’). As was expected, there was no deposition of actin5c-GFP in the entire nervous system because mef2Gal2 is expressed exclusively in the embryonic mesoderm (Fig. 1 F-F’’). We further examined the extracellular localization properties of Swim-GFP in late (stage 16) embryos, where somatic musculature is fully patterned, and compared Swim-GFP localization with actin5c-GFP. At this stage, actin filaments were deployed within somatic muscles and also heavily accumulated at the MAS, forming stable connections with integrins through the integrin adhesome machinery (Maartens and Brown, 2015). One of the extracellular matrix ligands of integrins that is localized at the MAS is tiggrin (Fogerty et al., 1994), (Fig. 2 A-A’). Similarly, Swim-GFP was highly accumulated at the MAS together with tiggrin, and was also found in the ventral midline colocalized with nidogen (Fig. 2 B-B’). Finally, we examined the distribution of Swim-GFP in late (stage 17) embryos (Fig. 2 C-E). On the dorsal side of the embryo, Swim-GFP was incorporated in the basement membrane surrounding the somatic muscles, and was also accumulated at the MAS and around the dorsal vessel (Fig. 2 C-C’). An optical section taken inside the embryo revealed that Swim-GFP was colocalized with nidogen in the basement membrane surrounding the entire gut, the somatic and pharyngeal muscles (Fig. 2 D-D’). On the ventral side of the embryo, Swim-GFP was tightly colocalized with nidogen in the ventral midline, while no longer decorating the neuropile (Fig. 2 E-E’).
Fig. 2. Swim-GFP is secreted from muscle cells and accumulates in tendon matrix and basement membranes in late Drosophila embryos.
(A-A’ and B-B’) Stage 16 and (C-E and C’-E’) stage 17 embryos probed with antibodies against nidogen (red) and tiggrin (blue) while expressing (A-A’) Actin5c-GFP (green) or (B-E and B’-E’) Swim-GFP (green) both driven by mef2Gal4. (A-A’) Lateral view of fully patterned somatic musculature as highlighted by Actin5c-GFP localization. Tiggrin accumulation decorates the stable connections of muscle attachment sites demonstrating the same (B-B’) Swim-GFP distribution pattern in a ventrolateral view of the somatic musculature. Swim-GFP colocalization with nidogen at the ventral midline persists at this stage of development. (C-C’) Superficial dorsal side view images reveal Swim-GFP accumulation at the muscle attachment sites along with its incorporation into the somatic muscle basement membrane and surrounding of the dorsal vessel. (D-D’) Internal dorsal side view images reveal colocalization of Swim-GFP with nidogen, both highlighting the basement membrane that encompasses the entire gut tissue, the somatic and pharyngeal muscles. (E-E’) Tight Swim-GFP accumulation at the ventral midline persists, as depicted by the superficial ventral side view. However, its distribution at the neuropile seems to have fallen off. Each image is representative of at least five different imaged embryos of the same genotype per staining: bm, basement membrane; dv, dorsal vessel; MAS, muscle attachment sites; vml, ventral midline. Scale bars: 50 μm in all panels.
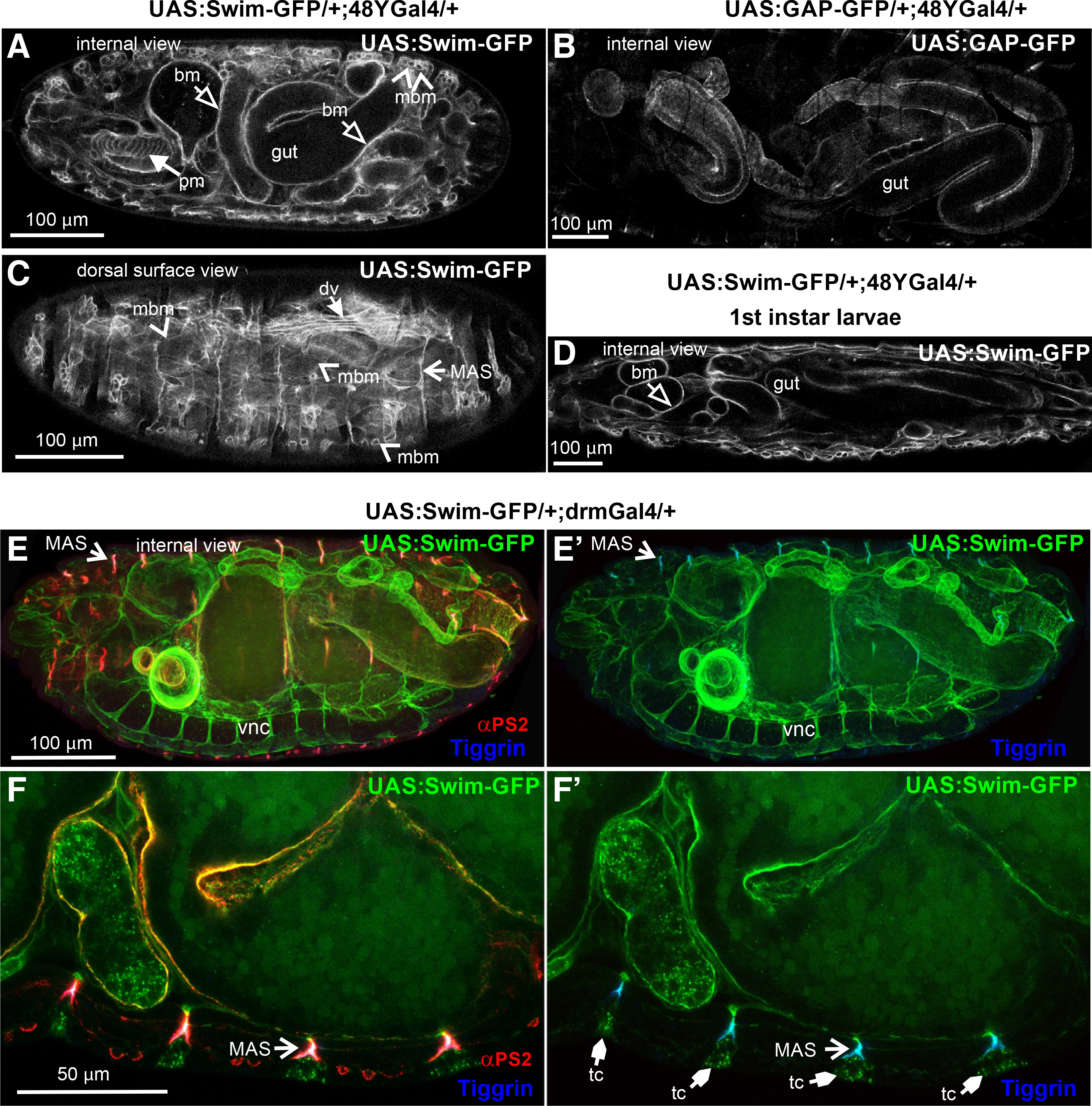
Secondly, we examined the extracellular distribution of Swim-GFP upon its expression in the 48YGal4-expressing cells in the gastrointestinal tract. We looked at late (stage 17) embryos and identified that Swim-GFP was detected not only in the areas encircling the gut but also in the pharyngeal and somatic muscles (Fig. 3A). By contrast, expression of GAP-GFP, which labels the plasma membrane, clearly outlined only the gut cells (Fig. 3B). Closer examination of late-stage embryos further revealed that Swim-GFP was incorporated into the basement membrane surrounding the somatic muscles, and was also enriched at the MAS, consistent with what we found with the mef2Gal4-mediated expression (Fig. 3C). Similar extracellular distribution of Swim-GFP was maintained at later developmental stages in larvae (Fig. 3D).
Fig. 3. Swim-GFP is secreted from epithelial cells and accumulates in basement membranes and tendon matrix in late Drosophila embryos.
(A-C) Stage 17 embryos expressing (A, C) Swim-GFP or (B) GAP-GFP both driven by 48YGal4. (A) Internal dorsal side view images reveal an accumulation of the Swim-GFP in the gut surroundings and in pharyngeal and somatic muscle tissues. However, neither of the latter seems to be denoted by (B) GAP-GFP localization used to highlight the 48YGal4 pattern of expression. Instead, a thorough examination of (C) Swim-GFP from a dorsal superficial view clearly outlines the protein enrichment in a repertoire of musculature tissue correlating with mef2Gal4 that persists (D) after completion of embryogenesis to the 1st instar larval stage. (E-F) Stage 17 embryos probed with antibodies against αPS2 integrin (red) and tiggrin (blue) while expressing Swim-GFP driven by drmGal4. (E-E’) Internal lateral side view images reveal the Swim-GFP distribution in multiple tissues including the ventral nerve cord, muscle attachment sites and the basement membrane surrounding the gut. (F-F’) Confocal cross section in high magnification depicting Swim-GFP accumulation at the tendon cells along with colocalization with tiggrin and αPS2 integrin subunit at the muscle attachment sites. Each image is representative of at least five different imaged embryos of the same genotype per staining: bm, basement membrane; dv, dorsal vessel; MAS, muscle attachment sites; mbm, muscle basement membrane; pm, pharyngeal muscles; tc, tendon cell; vnc, ventral nerve cord. Scale bars: 100 μm in panels A-E; 50 μm in panel F.
Thirdly, we expressed Swim-GFP with drmGal4, another Gal4 line that is expressed predominantly in the posterior part of the gut. We observed Swim-GFP distribution in regions of the ventral nerve cord, the basement membrane around the gut, and also at the MAS, colocalized with αPS2 integrin and tiggrin (Fig. 3 E-F). Collectively, our data indicate that Swim is secreted and deposited in the basement membranes of various organs, even if the tissue of its expression is not in proximity. For instance, Swim is secreted from the gut cells and enters the hemocoel cavity of the embryo and is gradually incorporated into the specialized tendon matrix at the MAS, or associated with the basement membranes surrounding various internal organs.
Swim-GFP accumulation in the tendon matrix does not depend on integrins
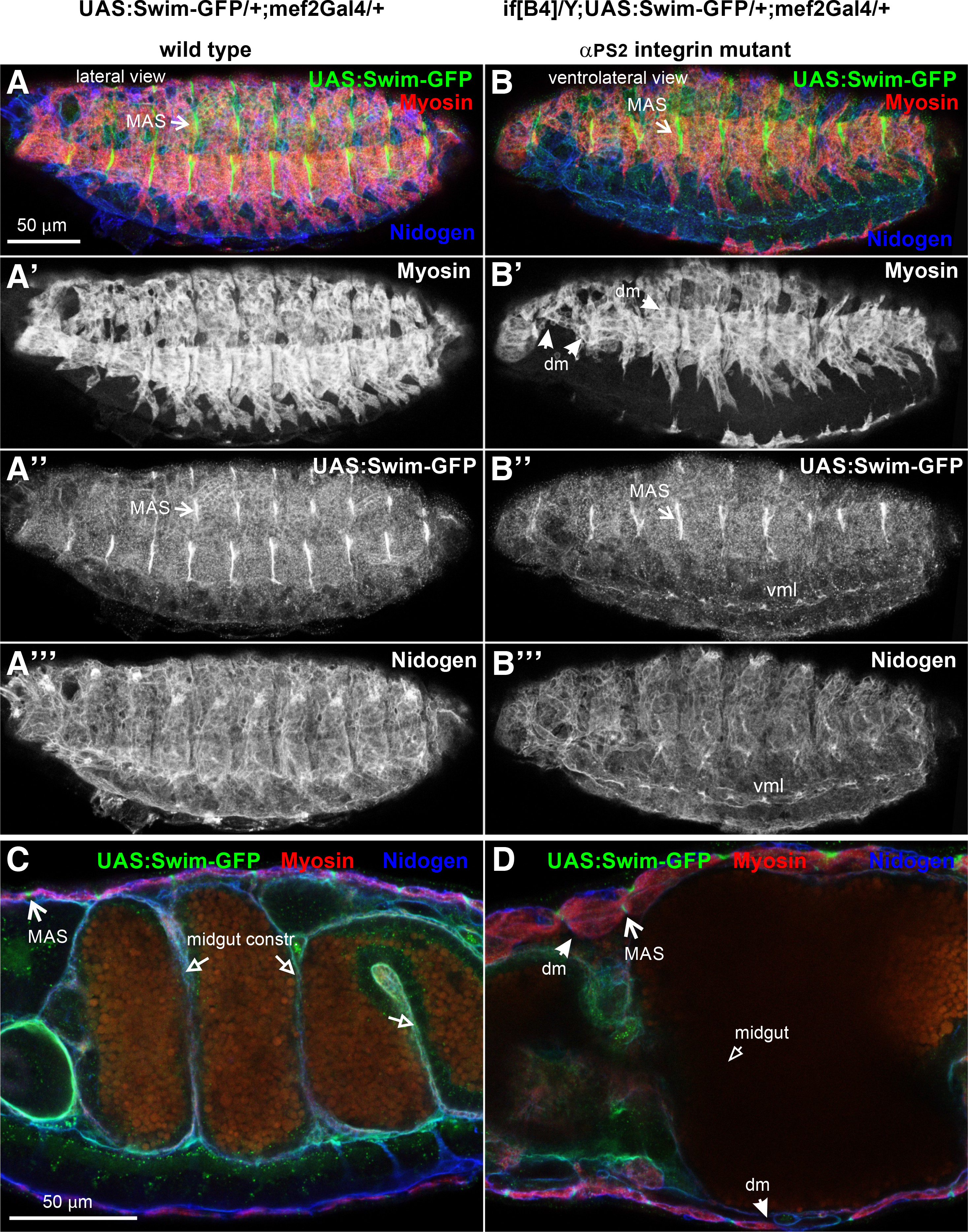
Earlier studies on TINAGL1 have shown its interaction with components of the basement membrane (Li et al., 2007). Interestingly, it was recently discovered that TINAGL1 was associated with α5β1 integrin in mouse LM2 cell line competing with fibronectin for integrin binding (Shen et al., 2019). Therefore, we examined whether the extracellular deposition of Swim-GFP depends on integrins in the Drosophila embryo. We utilized null inflated mutant embryos lacking the zygotic expression of αPS2 integrin, which is predominantly expressed in the somatic and visceral muscles (Brown, 1994). In the absence of αPS2 integrin, somatic muscles fail to maintain their adhesion to the tendon matrix and detach. Moreover, the midgut constrictions also fail to form in late embryos (stage 16) and the midgut acquires a sac-like morphology. We found no effect on the tight association of Swim-GFP at the MAS in the absence of integrins (Fig. 4 A-D), suggesting that Swim recruitment in the tendon matrix is mediated by other molecular cues. It will be interesting to examine whether Swim accumulation at the MAS or other basement membranes within the embryo is disrupted in absence of other ECM components (Urbano et al., 2009).
Fig. 4. Swim-GFP accumulation in the tendon matrix does not require integrin function.
Stage 16 (A-A’” and C) wild type and (B-B’” and D) null inflated (ifB4) mutant embryos probed with antibodies against myosin (red) and nidogen (blue) while expressing Swim-GFP (green) driven by mef2Gal4. (A-A’”) Lateral side view of wild type embryo denotes (A’) the somatic muscles with myosin (red in A and white in A”), (A”) and reveals the enrichment of Swim-GFP (green in A and white in A’’) at the muscle attachment sites and (A’”) the muscle basement membrane marked with nidogen. (B-B’”) Ventrolateral side view of an αPS2 mutant embryo. (B’) somatic muscles labelled with myosin, detach from the tendon matrix and form myospheres in the anterior and middle area of the embryo. (B”) Swim-GFP maintains its tight accumulation at the muscle attachment sites and (B’”) nidogen encompasses the muscle basement membrane. (C-D) Internal cross-section view of late (stage 16) embryo at high magnification. (C) in the wild type embryo the midgut constrictions have formed and Swim-GFP is enriched at MAS, whereas (D) in the αPS2 mutants, both the detached muscles and the characteristic failure in the formation of midgut constrictions are evident, with no effect on the robust accumulation of Swim-GFP at the tendon matrix. Each image is representative of at least five different imaged embryos of the same genotype per staining: dm, detached muscles; MAS, muscle attachment sites; vml, ventral midline. Scale bars: 50 μm in all panels.
Materials and Methods
Drosophila Genetics
All fly stocks and crosses were raised on standard cornmeal medium and kept at 25°C. To drive expression of the UAS: Swim-GFP or the UAS:Actin5c-GFP constructs (Verkhusha et al., 1999), the following GAL4 enhancer traps were obtained from Bloomington Stock Center: mef2-Gal4 (BL-50742), drmGal4 (BL-7098) and 48YGal4 (BL-4935) (FlyBaseConsortium, 1999).
Generation of UAS:Swim-GFP transgenic lines
A full-length cDNA for Swim was amplified from a 4-8 hr cDNA library using appropriate primers to construct the transgene (Brown and Kafatos, 1988). After confirmation by DNA sequencing, the coding region for Swim was subcloned in frame with a small four-serine linker followed by the mGFP6 that was derived from the Ilk-GFP construct (Zervas et al., 2001). Finally, the Swim-SSSS-GFP cassette was subcloned into the P-element transformation vector pUASp. Several transgenic lines were obtained, following standard protocols of embryos microinjection.
Immunofluorescence experiments and Microscopy
Embryos were collected from timed egg-lays at 25°C and staged appropriately as described (Campos-Ortega and Hartenstein, 1997). Whole embryo fixations were performed in 4% formaldehyde in PBS. PBT (0.5% BSA and 0.2% Triton X-100 in PBS or TBS respectively) was used for blocking, washes, primary and secondary antibody incubation either at room temperature or at 4°C. Primary antibodies used in this study were against MHC (myosin heavy chain, mouse monoclonal; 1:60) (Kiehart et al., 1990); anti-Tiggrin (gift of John Fessler, mouse pAb, 1:1000); anti-Nidogen (Wolfstetter et al., 2009); αPS2 integrin subunit (PS2HC2) (rat monoclonal; 1:10) (Bogaert et al., 1987).
All samples were mounted in Vectashield medium (Vector Laboratories). Single confocal sections and z stacks were acquired on a Leica TCS SP5 laser scanning inverted confocal microscope with an HC Plan Apochromat 20x/0.7 or HC Plan-Apochromat 63x/1.4 oil objective. Confocal settings were adjusted to avoid pixel intensity saturation of 1,024x1,024 pixel images captured at 400 Hz. Post-acquisition assembly was performed with LAS AF software (v.2.3.6).
Acknowledgements
This work was supported by AFMTéléthon (23004) and Fondation Santé to CGZ. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
Abbreviations
ECM, extracellular matrix ; GFP, green fluorescent protein ; MAS, muscle attachment sites ; Swim, secreted wingless-interacting protein ; TINAGL1, tubulointerstitial nephritis antigen like 1 ; UAS, upstream activating sequence ; Wg, wingless ;References
Bogaert T., Brown N., Wilcox M. (1987). The Drosophila PS2 antigen is an invertebrate integrin that, like the fibronectin receptor, becomes localized to muscle attachments. Cell 51: 929-940.
Bonnans C., Chou J., Werb Z. (2014). Remodelling the extracellular matrix in development and disease. Nature Reviews Molecular Cell Biology 15: 786-801.
Brand A.H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401-415.
Broadie K., Baumgartner S., Prokop A. (2011). Extracellular matrix and its receptors in drosophila neural development. Developmental Neurobiology 71: 1102-1130.
Brömme N. C., Wex T., Wex H., Levy B., Lipyansky A., Brömme D. (2000). Cloning, Characterization, and Expression of the Human TIN-ag-RP Gene Encoding a Novel Putative Extracellular Matrix Protein. Biochemical and Biophysical Research Communications 271: 474-480.
Brown N.H. (1994). Null mutations in the alpha PS2 and beta PS integrin subunit genes have distinct phenotypes. Development 120: 1221-1231.
Brown N. H., Kafatos F. C. (1988). Functional cDNA libraries from Drosophila embryos. Journal of Molecular Biology 203: 425-437.
Campos-Ortega J. A., Hartenstein V., (1997). The Embryonic Development of Drosophila melanogaster. Springer, Berlin, Heidelberg.
Walma D. A. C., Yamada K. M. (2020). The extracellular matrix in development. Development 147: dev175596.
Dai J., Estrada B., Jacobs S., Sánchez-Sánchez B. J., Tang J., Ma M., Magadán-Corpas P., Pastor-Pareja J. C., Martín-Bermudo M. D. (2018). Dissection of Nidogen function in Drosophila reveals tissue-specific mechanisms of basement membrane assembly. PLOS Genetics 14: e1007483.
Fakhouri M., Elalayli M., Sherling D., Hall J. D., Miller E., Sun X., Wells L., LeMosy E. K. (2006). Minor proteins and enzymes of the Drosophila eggshell matrix. Developmental Biology 293: 127-141.
FlyBase Consortium , (1999). The FlyBase Database of the Drosophila Genome Projects and community literature. Nucleic Acids Research 27: 85-88.
Fogerty F.J., Fessler L.I., Bunch T.A., Yaron Y., Parker C.G., Nelson R.E., Brower D.L., Gullberg D., Fessler J.H. (1994). Tiggrin, a novel Drosophila extracellular matrix protein that functions as a ligand for Drosophila alpha PS2 beta PS integrins. Development 120: 1747-1758.
Hynes R. O. (2009). The Extracellular Matrix: Not Just Pretty Fibrils. Science 326: 1216-1219.
Hynes R. O., Naba A. (2012). Overview of the Matrisome--An Inventory of Extracellular Matrix Constituents and Functions. Cold Spring Harbor Perspectives in Biology 4: a004903-a004903.
Igarashi T., Tajiri Y., Sakurai M., Sato E., Li D., Mukai K., Suematsu M., Fukui E., Yoshizawa M., Matsumoto H. (2009). Tubulointerstitial Nephritis Antigen-Like 1 Is Expressed in Extraembryonic Tissues and Interacts with Laminin 1 in the Reichert Membrane at Postimplantation in the Mouse1. Biology of Reproduction 81: 948-955.
Kalfa T.A., Thull J.D., Butkowski R.J., Charonis A.S. (1994). Tubulointerstitial nephritis antigen interacts with laminin and type IV collagen and promotes cell adhesion.. Journal of Biological Chemistry 269: 1654-1659.
Kiehart D. P., Ketchum A., Young P., Lutz D., Alfenito M. R., Chang X., Awobuluyi M., Pesacreta T. C., Inoué S., Stewart C. T., Chen T.L. (1990). Contractile Proteins in Drosophila Development. Annals of the New York Academy of Sciences 582: 233-251.
Korpal M., Ell B. J., Buffa F. M., Ibrahim T., Blanco M. A., Celià-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y., Wei Y., Hu G., Garcia B. A., Ragoussis J., Amadori D., Harris A. L., Kang Y. (2011). Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nature Medicine 17: 1101-1108.
Maartens A. P., Brown N. H. (2015). The many faces of cell adhesion during Drosophila muscle development. Developmental Biology 401: 62-74.
McGough I. J., Vecchia L., Bishop B., Malinauskas T., Beckett K., Joshi D., O’Reilly N., Siebold C., Jones E. Y., Vincent J.P. (2020). Glypicans shield the Wnt lipid moiety to enable signalling at a distance. Nature 585: 85-90.
Mulligan K. A., Fuerer C., Ching W., Fish M., Willert K., Nusse R. (2012). Secreted Wingless-interacting molecule (Swim) promotes long-range signaling by maintaining Wingless solubility. Proceedings of the National Academy of Sciences 109: 370-377.
Musetti S. N., Huang L. (2021). Tinagl1 Gene Therapy Suppresses Growth and Remodels the Microenvironment of Triple Negative Breast Cancer. Molecular Pharmaceutics 18: 2032-2038.
Sakurai M., Sato Y., Mukai K., Suematsu M., Fukui E., Yoshizawa M., Tanemura K., Hoshino Y., Matsumoto H., Sato E. (2014). Distribution of tubulointerstitial nephritis antigen-like 1 and structural matrix proteins in mouse embryos during preimplantation development in vivo and in vitro . Zygote 22: 259-265.
Shen M., Jiang Y.Z., Wei Y., Ell B., Sheng X., Esposito M., Kang J., Hang X., Zheng H., Rowicki M., Zhang L., Shih W. J., Celià-Terrassa T., Liu Y., Cristea I.I., Shao Z.M., Kang Y. (2019). Tinagl1 Suppresses Triple-Negative Breast Cancer Progression and Metastasis by Simultaneously Inhibiting Integrin/FAK and EGFR Signaling. Cancer Cell 35: 64-80.e7.
Takahashi A., Rahim A., Takeuchi M., Fukui E., Yoshizawa M., Mukai K., Suematsu M., Hasuwa H., Okabe M., Matsumoto H. (2016). Impaired female fertility in tubulointerstitial antigen-like 1-deficient mice. Journal of Reproduction and Development 62: 43-49.
Urbano J. M., Torgler C. N., Molnar C., Tepass U., López-Varea A., Brown N. H., de Celis J. F., Martín-Bermudo M. D. (2009). Drosophila laminins act as key regulators of basement membrane assembly and morphogenesis . Development 136: 4165-4176.
Verkhusha V. V., Tsukita S., Oda H. (1999). Actin dynamics in lamellipodia of migrating border cells in the Drosophila ovary revealed by a GFP-actin fusion protein . FEBS Letters 445: 395-401.
Wex T., Lipyansky A., Brömme N. C., Wex H., Guan X. Q., Brömme D. (2001). TIN-ag-RP, a Novel Catalytically Inactive Cathepsin B-Related Protein with EGF Domains, Is Predominantly Expressed in Vascular Smooth Muscle Cells , . Biochemistry 40: 1350-1357.
Wolfstetter G., Dahlitz I., Pfeifer K., Töpfer U., Alt J. A., Pfeifer D. C., Lakes-Harlan R., Baumgartner S., Palmer R. H., Holz A. (2019). Characterization of Drosophila Nidogen / entactin reveals roles in basement membrane stability, barrier function and nervous system patterning . Development 146: dev168948.
Wolfstetter G., Shirinian M., Stute C., Grabbe C., Hummel T., Baumgartner S., Palmer R. H., Holz A. (2009). Fusion of circular and longitudinal muscles in Drosophila is independent of the endoderm but further visceral muscle differentiation requires a close contact between mesoderm and endoderm. Mechanisms of Development 126: 721-736.
Zervas C. G., Gregory S. L., Brown N. H. (2001). Drosophila Integrin-Linked Kinase Is Required at Sites of Integrin Adhesion to Link the Cytoskeleton to the Plasma Membrane. Journal of Cell Biology 152: 1007-1018.